Information
- Author Services

Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu
- Subscribe SciFeed
- PubMed/Medline
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Plants: an international scientific open access journal to publish all facets of plants, their functions and interactions with the environment and other living organisms.

Introduction
Plants as a source of food:, the environmental impact:, biodiversity:, references and notes.
- Bennett, B.C. Plants as Food. Bennett, Brad, Ed.; Developed under the Auspices of the UNESCO; Eolss Publishers: Oxford, UK, 2010. [ Google Scholar ]
- Zeenews.com. Available online: http://zeenews.india.com/sci-tech/ecology/index26.html (accessed on 25 January 2012).
- Planetsave. Available online: http://planetsave.com/2011/05/20/endangered-plants-list/ (accessed on 16 January 2012).
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fernando, W.G.D. Plants: An International Scientific Open Access Journal to Publish All Facets of Plants, Their Functions and Interactions with the Environment and Other Living Organisms. Plants 2012 , 1 , 1-5. https://doi.org/10.3390/plants1010001
Fernando WGD. Plants: An International Scientific Open Access Journal to Publish All Facets of Plants, Their Functions and Interactions with the Environment and Other Living Organisms. Plants . 2012; 1(1):1-5. https://doi.org/10.3390/plants1010001
Fernando, W.G. Dilantha. 2012. " Plants: An International Scientific Open Access Journal to Publish All Facets of Plants, Their Functions and Interactions with the Environment and Other Living Organisms" Plants 1, no. 1: 1-5. https://doi.org/10.3390/plants1010001
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Review Article
- Open access
- Published: 01 April 2021
Research advances in and prospects of ornamental plant genomics
- Tangchun Zheng 1 ,
- Ping Li 1 ,
- Lulu Li 1 &
- Qixiang Zhang 1
Horticulture Research volume 8 , Article number: 65 ( 2021 ) Cite this article
8781 Accesses
37 Citations
3 Altmetric
Metrics details
- DNA sequencing
- Plant genetics
The term ‘ornamental plant’ refers to all plants with ornamental value, which generally have beautiful flowers or special plant architectures. China is rich in ornamental plant resources and known as the “mother of gardens”. Genomics is the science of studying genomes and is useful for carrying out research on genome evolution, genomic variations, gene regulation, and important biological mechanisms based on detailed genome sequence information. Due to the diversity of ornamental plants and high sequencing costs, the progress of genome research on ornamental plants has been slow for a long time. With the emergence of new sequencing technologies and a reduction in costs since the whole-genome sequencing of the first ornamental plant ( Prunus mume ) was completed in 2012, whole-genome sequencing of more than 69 ornamental plants has been completed in <10 years. In this review, whole-genome sequencing and resequencing of ornamental plants will be discussed. We provide analysis with regard to basic data from whole-genome studies of important ornamental plants, the regulation of important ornamental traits, and application prospects.
Similar content being viewed by others

Chromosome-level genomes of three key Allium crops and their trait evolution

Chromosome-level genome assembly, annotation and evolutionary analysis of the ornamental plant Asparagus setaceus
Combination of long-read and short-read sequencing provides comprehensive transcriptome and new insight for Chrysanthemum morifolium ray-floret colorization
Introduction.
Genomics is the science of studying genomes. It is used to summarize a branch of genetics involving genome mapping, sequencing, and whole-genome functional analysis. The whole genome is taken as the research object, with a focus on analyzing all of the genetic information in whole genomes of organisms. The main purpose of carrying out genomics research is to interpret the whole-genome sequence, including genomic variations and gene regulation, through mining and expression to gain a deeper understanding of biological mechanisms, formulate more effective breeding strategies, expand the mining breadth and depth of excellent alleles in germplasm resources, and increase the operability for improving complex traits and the efficiency of breeding new varieties.
Ornamental plants, a vital component of agriculture and horticulture, are of great significance for beautifying and improving humans’ living environment, cultivating human sentiment, and promoting structural adjustments in the agricultural industry. The first plant genome to be published was that of Arabidopsis thaliana in 2000 1 . With the emergence of next-generation and high-throughput sequencing, sequencing technologies have continuously evolved, while their costs have continuously decreased, facilitating the whole-genome sequencing of many plants. According to incomplete statistics, whole-genome sequencing has been completed for ~400 plants 2 . With this progress, more abundant genetic data are provided for plant diversity studies, enabling breeders to perform comprehensive multidimensional research in the fields of genetics, genomics, and molecular breeding. This brings new development opportunities and driving forces for the breeding of more plants and thus leads to a new revolution of breeding technology. Since genome sequencing of the first ornamental plant ( Prunus mume ) was completed in 2012 3 , whole-genome sequencing of more than 65 ornamental plants has been completed in <10 years. The whole-genome sequencing results from these ornamental plant species have built an enormous resource platform for molecular biology research in ornamental horticulture, which not only contributes to the understanding of genome structure and function in ornamental horticulture but also has substantial guiding significance for exploring the origin and evolution of ornamental plants, mapping and cloning the functional genes of important traits and accelerating the course of molecular breeding.
In this study, the research results from whole-genome sequencing and resequencing of ornamental plants are summarized. We provide a discussion with regard to basic data from whole-genome studies of important ornamental plants, the regulation of important ornamental traits, and application prospects.
Whole-genome sequences of ornamental plants
As of 30 October 2020, the whole-genome sequences and draft genome sequences of 69 ornamental plants have been published, including herbaceous plants, such as carnation ( Dianthus caryophyllus ), phalaenopsis ( Phalaenopsis aphrodite ), orchid ( Apostasia odorata ), sacred lotus ( Nelumbo nucifera ), chrysanthemum ( Dendranthema morifolium ) and Dionaea muscipula , and woody plants, such as mei ( Prunus mume ), Yoshino cherry ( Prunus yedoensis ), sweet osmanthus ( Osmanthus fragrans ), peony ( Paeonia suffruticosa ), and Chinese rose ( Rosa chinensis ) (Table 1 ). The number of sequenced genomes of ornamental plants completed each year significantly increased from 1 in 2012 to 17 in 2018. In particular, more than 10 species were sequenced for three consecutive years from 2016 to 2018 (Fig. 1a ). China has independently completed or led genome sequencing for 32 ornamental plants, followed by Japan and the United States, which have also completed the genome sequencing of more than 10 species (Fig. 1b ). Considering the sequencing material, except for the double-haploid material with relatively high homozygosity used for R. chinensis 4 , 5 , wild diploids or cultivars with relatively unclear genetic backgrounds and low heterozygosity were used for all of the other plants. Long-read sequencers in combination with optical maps 6 are used to generate high-quality chromosome-level genome assemblies. For ornamental plants, the PacBio RS II system was first applied for the construction of the 1.27 Gb genome assembly of Dendrobium officinale 7 . Long-range scaffolding techniques such as high-throughput chromosome conformation capture (Hi-C) facilitate chromosome-scale assembly of contigs. In this respect, recently built genome assemblies of Rosa chinensis (515 Mb) have a contig N50 of 24 Mb, which is one of the most comprehensive plant genomes 4 . In consideration of the comprehensive utilization of Illumina HiSeq, Nanopore, PacBio, and Hi-C technologies, the contig N50 values of Gardenia jasminoides and Chimonanthus praecox can reach 44 and 65.35 Mb, respectively, which was unthinkable five years ago 8 , 9 . Generally, the sequencing technology that is predominantly used is next-generation sequencing on the Illumina platform (HiSeq 2000/2500/4000 and HiSeq X ten), coupled with third-generation sequencing (PacBio and Nanopore) and Hi-C technology. The assembled genome size of sequenced ornamental plants ranges from 237 Mb to 13.79 Gb with a scaffold N50 ranging from 13.8 Kb to 65.35 Mb (Fig. 2 ). We constructed phylogenetic trees for all species with a published genome, which belong to 21 orders and 35 families (Fig. 3 ). The representative species in Rosaceae, Orchidaceae, and Asteraceae for which high-quality sequencing has been completed were described and discussed.
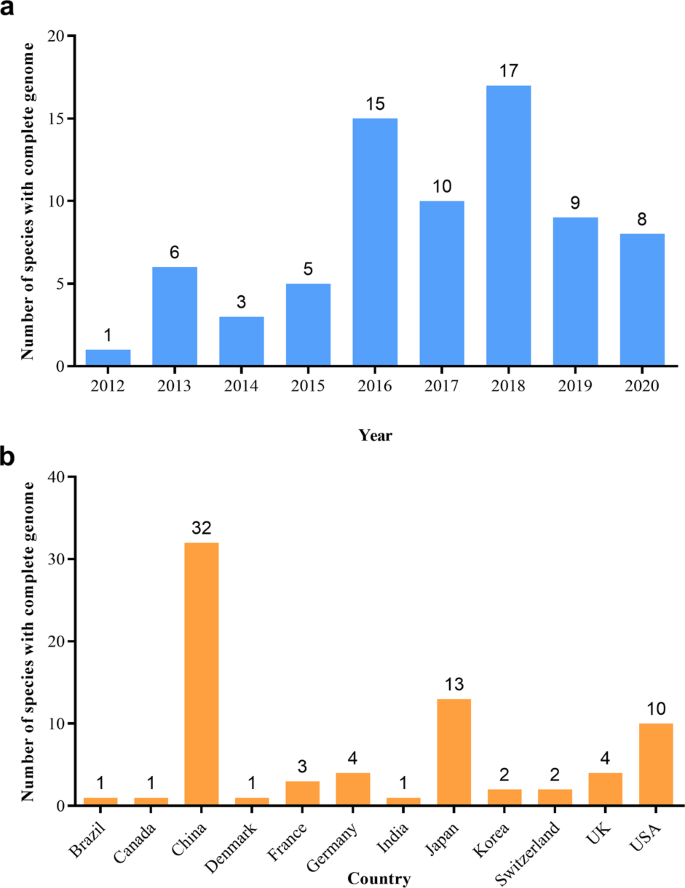
a Distribution of genome sequencing for ornamental plants completed from 2012 to 2020; b Distribution of genome sequencing for ornamental plants completed in different countries
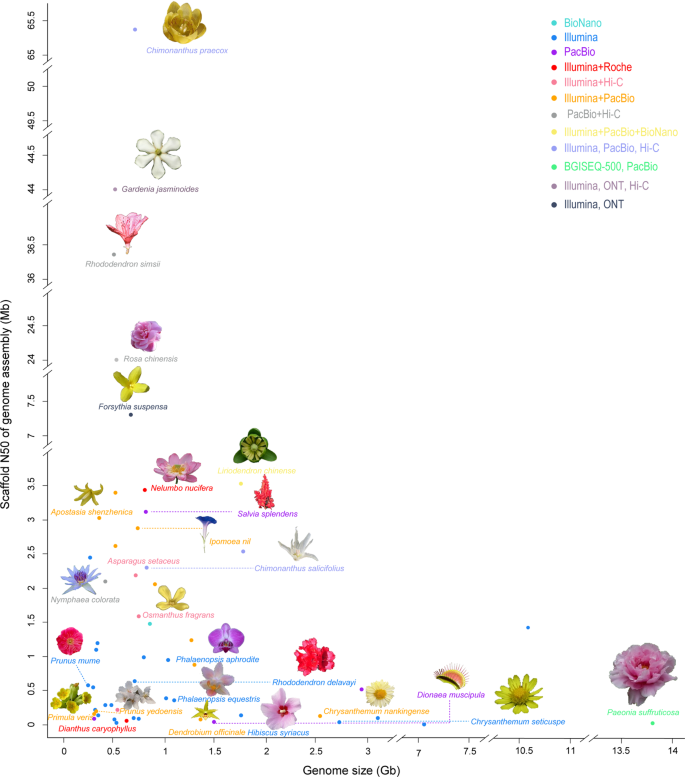
The x -axis represents the genome size of each plant, while the y -axis shows the scaffold N50 of the genome assembly. The sequencing platforms are indicated in different colors
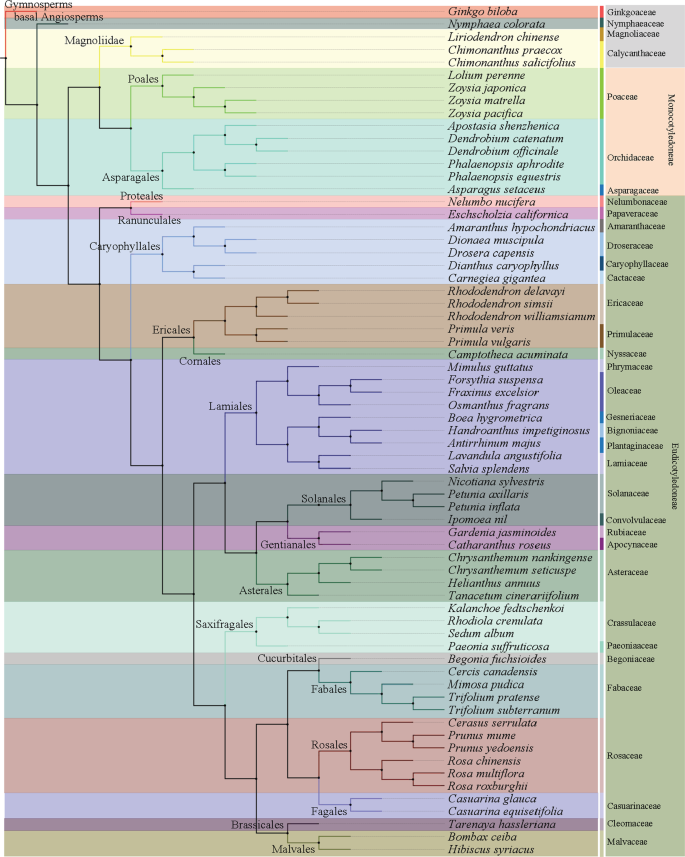
A maximum likelihood (ML) phylogenetic tree was built using low-copy orthologous sequences. All the published ornamental species belong to 21 orders and 35 families. The same background color was used for species in the same family
Rosaceae contains more than 3300 species in 124 genera that are rich in economic and ornamental value and occupy an important position in gardens worldwide. The first flowering ornamental plant to be sequenced was Prunus mume (mei) from Rosaceae. In 2009, the National Engineering Research Center for Floriculture of Beijing Forestry University cooperated with the Beijing Genomics Institute (BGI) and other institutions to launch the mei genome project. First, a 237 Mb (84.6% of the estimated genome) genome of wild-type mei was assembled using the Illumina GA II. The scaffold N50 was 577.8 Kb, and 31,390 protein-coding genes were annotated. The genome data were published in Nature Communications in 2012, and this effort marked the first genome sequence map of a flowering crop worldwide 3 . Interestingly, equal to the status of mei in China, the “Yoshino cherry” tree ( Prunus × yedoensis ) is one of the most popular Prunus species in Japan, and its genome was sequenced by Korean researchers, revealing the parental origin and genomic delimitation of hybrid taxa using both Illumina and PacBio platforms in 2018 10 . Soon afterwards, researchers from Japan also completed two similar genomes of Cerasus yedoensis , “Somei-Yoshino”, which were merged into a special genome 11 . At present, a large number of genome studies focusing on Prunus and Rosa in Rosaceae are underway.
Roses have high cultural and economic value as the most commonly cultivated ornamental and spice plants worldwide. The first ornamental Rosa to have its genome sequenced was Rosa multiflora , which was reported by Japanese scholars focusing on flower color, flower scent, and floral development traits 12 . Then, another well-known and long-awaited major study was published in Nature Genetics in May 2018. A team at the University of Lyon and Centre National de la Recherche Scientifique (CNRS) first revealed another parent of the modern rose, Rosa chinensis . The size of the Rosa genome is 560 Mb with a contig N50 of 24 Mb, which is one of the most comprehensive plant genomes 4 . Coincidentally, one month later, the same experimental material (a doubled haploid line from ‘Old Blush’) of Rosa chinensis was sequenced and republished in Nature Plants in June 2018. The high-quality genome was cross-verified, and ornamental and production traits of rose have been interpreted with the joint efforts of many research institutions from France, Belgium, Russia, etc. 5 .
Orchidaceae
As one of the most abundant families in the plant kingdom, Orchidaceae (orchid) plants are the flagship species of plant diversity protection, known as the “panda of the plant kingdom”. Orchids are divided into five subfamilies: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae. Phalaenopsis and Dendrobium belong to Orchidoideae and Epidendroideae. Phalaenopsis plants are representative of Orchidaceae plants and have important ornamental value. Professor Zhongjian Liu of the National Orchid Conservation Center of China overcame technical problems resulting from high heterozygosity and completed the assembly of the whole-genome sequence of P. equestris with a scaffold N50 size of 359.1 Kb. As the first monocot flower for which genome-wide sequencing was completed, the genome of P. equestris was published as a cover paper in the journal Nature Genetics in November 2014 13 . Phalaenopsis is an important potted flower with high economic value worldwide. A 3.1 Gb draft genome assembly of an important winter-flowering Phalaenopsis cultivar ‘KHM190’ was completed by researchers from China and Australia 14 . Another species of Phalaenopsis , P. aphrodite , also underwent high-quality genome sequencing with a scaffold N50 size of 19.7 Mb in April 2018 15 . Scholars from China further analyzed the whole genomes of Dendrobium officinale and Dendrobium catenatuma , which were published in the journals Molecular Plant and Scientific Reports , respectively 7 , 16 . Apostasia shenzhenica is representative of one of two genera that form a sister lineage with the rest of the Orchidaceae ; they have unique flower morphologies as well as diverse lifestyles and habitats. Professor Zhongjian Liu resequenced the high-quality genome of A. shenzhenica with a scaffold N50 size of 3.0 Mb. A 349 Mb genome was assembled and published in Nature in 2017 17 . Vanilla fragrans is a plant of the vanilla family. Due to its unique fragrance that cannot be synthesized artificially, it is known as the “Perfume Queen”. In July 2014, the Fujian Agriculture & Forestry University and National Orchid Conservation Center of China (Shenzhen) officially launched the Vanilla shenzhenica genome project. As the first Orchidaceae vine plant to undergo complete sequencing, the genome of V. shenzhenica was ~800 Mb with a scaffold N50 size of 288 Kb, and its heterozygosity was ~1.14% ( https://www.fafu.edu.cn/2015/0208/c132a18466/page.htm ).
There are ~24,000–35,000 species in Asteraceae; this family has very high plant diversity, accounting for ~10% of total angiosperms. Chrysanthemum , as a typical representative genus, is one of the most important ornamental crops in the world. The genome of Chrysanthemum morifolium is estimated to be more than 9 Gb ( http://data.kew.org/cvalues/ ). Since the Chrysanthemum genus is large and complex, the genome of Chrysanthemum was not reported for a long time. In October 2018, the China Academy of Chinese Medical Sciences, Hubei University of Chinese Medicine cooperated with Nanjing Agricultural University and completed the sequencing of Chrysanthemum nankingense , a diploid species (2 n = 18), which represents one of the progenitor genomes of domesticated chrysanthemums 18 . At around the same time, the de novo whole-genome assembly of Chrysanthemum seticuspe was announced by researchers from the Kazusa DNA Research Institute of Japan 19 . The 2.72 Gb of assembled sequences covered 89.0% of the 3.06 Gb C. seticuspe genome with 71,057 annotated genes 19 . Sunflower ( Helianthus annuus L.), in the Asteraceae and the Helianthus genus, is a horticultural crop with important economic and ornamental value and a major research focus. In May 2017, a high-quality reference for the sunflower genome was published in the journal Nature by scientists from France and Canada 20 . The size of the sunflower genome was 2.94 Gb and covered 80% of the estimated genome; finally, 97% of annotated genes were anchored on a total of 17 pseudochromosomes.
Resequencing of ornamental plants
Whole-genome resequencing is a process of sequencing the genomes of different individuals of species with known genome sequences and analyzing the differences among individuals or populations. In recent years, to overcome the narrow genetic variation in current ornamental plant breeding programs, genome-scale investigations of wide germplasm panels and cultivated varieties have served to identify important genetic materials to study genomic variation dynamics during domestication and selective breeding 71 . For example, resequencing of multiple materials from different crop species based on genome-wide association study (GWAS) was facilitated to identify key genomic regions associated with plant domestication and selection/improvement 72 . Based on genome-wide resequencing technology, researchers can quickly screen resources, find a large number of genetic variations, and realize genetic evolution analysis and prediction of important candidate genes. Although great progress has been made in the de novo sequencing of ornamental plant genomes, only a few species of ornamental plants, such as sunflower, lotus, mei, rose, sakura, and Liriodendron chinense , have undergone genome resequencing (Table 2 ).
Sunflower is not only an ornamental plant but also one of the four major oil crops in the world. In June 2017, genome sequencing of sunflower was completed, eighty domesticated lines (10–20× coverage) and 72 inbred lines (9.3–19.5× coverage) from 480 F 1 hybrids were resequenced, and 35 genomic regions associated with flowering time were identified by GWAS 20 . Subsequently, to characterize genetic diversity in sunflower and to quantify contributions from wild relatives, scientists from the University of British Columbia sequenced 493 accessions, including cultivars, landraces, and wild relatives 73 . In all, 61,205 genes have been identified within the gene set of the sunflower pangenome, and a large number of candidate resistance genes and single nucleotide polymorphism (SNP) markers for downy mildew resistance were identified by GWAS, which may be of interest to other researchers and sunflower breeders 73 .
To reveal the evolutionary history of Prunus mume and the Prunus genus and the genetic mechanism of important ornamental characteristics of P. mume , 333 cultivated landraces, 15 wild P. mume , and three close relatives of Prunus ( P. sibirica , P. davidiana , and P. salicina ) were selected for genome-wide resequencing by Professor Qixiang Zhang from the National Engineering Research Center for Floriculture of China 74 . A total of 5.34 million high-quality SNPs were identified, and 24 important ornamental traits (such as petal color, stigma color, calyx color, bud color, stamina filament color, wood color, petal number, pistil character, bud aperture, and branching phenotype) of 333 cultivars of P. mume were analyzed by GWAS for the first time to confirm the hypothesis that P. mume exists due to introgression from P. sibirica and P. salicina 74 .
Three versions of the lotus genome have been published in five years 21 , 24 , 50 . To explore the genomic diversity and microevolution related to the rhizome growth pattern, especially the genomic markers of ecotype differentiation, researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences resequenced 19 individuals including rhizome lotus, seed lotus, flower lotus, wild lotus, Thai lotus and Nelumbo lutea 75 . Candidate genes associated with temperate and tropical lotus divergence always exhibited highly divergent expression patterns, which are valuable for the breeding and cultivation of lotus 75 .
Roses have high cultural and economic value because of their outstanding ornamental characteristics and essential oil composition. To analyze the genetic diversity and genetic regulation mechanism of important ornamental traits in roses, eight Rosa species representing three of the four subgenera ( R. persica , R. minutifolia and Rosa ) were resequenced, and the whole-genome sequence of a double-haploid rose line was completed 5 . At the same time, to gain insight into the makeup of modern roses, Raymond et al. 4 resequenced representatives of three sections (“Synstylae”, “Chinenses” and “Cinnamomeae”) that participated in the domestication and breeding of the modern hybrid rose after the genome of homozygous Rosa chinensis ‘Old Blush’ was sequenced.
Sakura ( Prunus yedoensis ) is a woody ornamental plant with important cultural and economic value. To study the genomic relationship between P. yedoensis and its closely related species, nine P. yedoensis accessions and seven accessions of candidate parental species, including P. pendula , P. jamasakura and P. sargentii , were resequenced and compared to the assembled genome by researchers from Korea 10 . Resequencing data of six related taxa show that 41% of the genes were assigned to the parent state, suggesting that wild P. yedoensis is an F 1 hybrid originating from a cross between P. pendula and P. jamasakura 10 .
Liriodendron chinense is an important woody ornamental plant known as a “woody tulip” in the UK and USA, as its flower shape is similar to that of the tulip. The high-quality genome of L. chinense was published in the journal Nature Plants in December 2018 in a project led by Professor Jisen Shi from Nanjing Forestry University 57 . To explore the historical demographic fluctuations and present-day genetic diversity between L. chinense and L. tulipifera , 14 L. chinense individuals and 6 L. tulipifera individuals were resequenced. Population analysis showed that Liriodendron can be divided into three subgroups: the Eastern China subgroup, Western China subgroup and North American subgroup. The species divergence time confirmed that the genetic diversity of L. chinense was much higher than that of L. tulipifera 57 .
Applications of whole-genome sequencing in ornamental plants
Gene annotation.
Gene annotation is the process of attributing biological information to the completed sequence of a species using bioinformatics methods. It identifies gene fragments that do not encode proteins, recognizes elements on genes (gene prediction) and adds biological information to the elements for sequence repeat identification, noncoding RNA prediction, gene structure prediction, and gene function annotation. In this way, genes associated with ornamental horticultural traits such as flowering regulation, flower color, floral fragrance, plant type, dormancy, cold resistance, and disease resistance can be identified. The dormancy-associated MADS-box transcription factor (DAM) family, which is related to dormancy induction and release, is especially critical for ornamental plants 76 . Zhang et al. 3 identified six DAM genes in the tandem array in the P. mume genome and confirmed that the distribution pattern was consistent with that from previous studies of the peach genome 77 . In Rosa , Raymond et al. 4 identified new candidate genes potentially involved in recurrent blooming, such as TFL1 , SPT , and DOG1 .
Comparative genomics research
Based on genome mapping and sequencing technologies, comparative genomics research compares known genes and genome structures to understand the functions of associated genes, their expression mechanism, and the phylogenetic relationships of species. The acquisition of genomic information from multiple closely related species facilitates more comprehensive and in-depth research in comparative genomics. Moreover, it is crucial to perform in-depth comparative analysis of the collinear relationship between the genome sequences of two plants to analyze the origin and evolutionary relationship of plants and to explore important chromosome fragments or gene clusters that control major plant traits, which can provide essential reference information for the discovery and cloning of important genes. Zhang et al. constructed nine ancestral chromosomes of the Rosaceae family by comparing Rosaceae genomes. For the first time, these researchers revealed that ancestral chromosomes have evolved into eight existing chromosomes in P. mume via 11 fusions, seven existing chromosomes in strawberry ( Fragaria ananassa ) via 15 fusions and 17 existing chromosomes in apple ( Malus domestica ) via one whole-genome duplication event plus five fusions. These findings lay an important foundation for research to unravel the origin and evolution of Rosaceae 3 .
Resequencing
Whole-genome resequencing involves the sequencing of genomes in different individuals of species with known genome sequences and subsequent analysis of differences among individuals or populations. Whole-genome resequencing technology can be used to rapidly conduct resource screening, to find a large number of genetic variations and to implement genetic evolution analysis and candidate gene prediction for important traits. These results provide essential references for identifying valuable genetic resources and for horticultural crop breeding and are thus of significant research and industrial value. In P. mume , researchers investigated the genetic architecture of floral traits and plant domestication history by resequencing 348 P. mume accessions and three other Prunus species at an average sequencing depth of 19.3×. Highly admixed population structure and introgression from Prunus species were identified in mei accessions 74 . Huang et al. 75 resequenced and analyzed the genomes of 19 lotus germplasms, provided a reliable and detailed understanding of the genome evolution of different lotus germplasms, and provided clues to key mutations responsible for rhizome enlargement.
A GWAS is a genome-wide comparative analysis or correlation analysis using millions of SNPs in the genome as molecular genetic markers. It is a new strategy to find genetic variations that affect complex traits by comparison. With the development of genomics research and DNA microarray technology, a GWAS can provide an outlined overview of important traits simultaneously and is therefore suitable for the study of complex traits. At the genome-wide level, association studies between genes and traits are conducted with multiple centers, large samples, and repeated verifications. This method has been applied for the screening and identification of major genes for important economic traits in agriculture. In P. mume , through a GWAS, researchers have identified significant quantitative trait loci (QTLs) and genomic regions where several genes associated with petal color, stigma color, calyx color, bud color, stamina filament color, wood color, petal number, pistil character, bud aperture, and branching phenotype are located 74 . Taken together, the identification of genetic loci associated with floral and other traits provides more insight into the genetic mechanisms that underlie the domestication of P. mume and provides opportunities to design strategies for genomic selection to improve the performance of ornamental species. In sunflowers and roses, the key ornamental trait of flowering time was also identified by the GWAS method 4 , 20 .
Comparative analysis with transcriptome data
RNA sequencing is a newly emerging technology that uses next-generation sequencing for transcriptome analysis. It can comprehensively and rapidly acquire sequence information and expression information for almost all transcripts from specific cells or tissues in a particular state, including protein-coding mRNAs and various noncoding RNAs, as well as the expression abundance of different transcripts generated by alternative gene splicing. The transcriptome is an inevitable link that connects genetic information of the genome with the biological functions of the proteome. Currently, transcriptional regulation is the most well-studied and foremost regulatory method in organisms. Transcriptome studies are the foundation and starting point of gene function-structure studies and the first issue to address after the completion of whole-genome sequencing. Furthermore, transcriptome analysis provides large numbers of molecular markers, such as simple sequence repeats and SNPs. All of the sequence information, expression data, and molecular markers facilitate the localization of QTLs for key ornamental traits in ornamental plants through genetic mapping and contribute to the development of molecular markers in close linkage with excellent traits for use in the molecular marker-assisted breeding of flowers. Based on the genome sequence of P. mume , vital differences in gene expression between the bud stage and squaring stage were observed, and 7,813 DEGs were identified, which provided a special perspective on floral scent formation in P. mume 78 . The water lily genome revealed variable genomic signatures of ancient vascular cambium losses, and the expression profiles of floral ABCE genes, floral scent and color genes were screened from the DEGs in a comparative analysis of the transcriptome 64 .
Development of SNP microarrays
According to their position in genes, SNPs can occur in coding regions, noncoding regions, and gene spacer regions. They are DNA molecular markers that have the most abundant polymorphisms in the genome and are characterized by large numbers, a uniform distribution, and easy typing. SNPs can be used for the identification of genetic variation and genotyping of associated phenotypes. Using SNPs as molecular markers to construct genetic variation maps of the genome has become a vital part of the research for studying genome diversity, obtaining domesticated selection regions, and screening key genes of important traits. Based on the genome sequence and resequencing of P. mume , a total of 1,298,196 raw SNPs were located within coding regions of genes, 733,292 of which were nonsynonymous 74 . Furthermore, by combining transcriptome data, 76 SNPs within DEGs were identified that were associated with petal, stigma, calyx, and bud color 74 . In sacred lotus , wild and Thai lotus exhibited greater differentiation with a higher genomic diversity than cultivated lotus based on SNP sites in resequenced species 75 .
Exploiting genes associated with important ornamental traits
During the course of whole-genome sequencing, a very large number of genes, in the range of 19,507–87,603, are annotated for each flowering species (Table 1 ). Through further analysis, important genes associated with floral development, flower color formation, and stress resistance can be discovered. This is conducive to the breeding of unique, high-quality, and high-resistance varieties or types of a species and provides important references for improving ornamental and resistance qualities in other flowering species.
Candidate genes for controlling floral development
Flower blooming is a process that involves the formation of inflorescence meristems and flower meristem tissues through floral induction and a series of internal and external factors, followed by the generation of floral organ primordia and eventually the release of flora bud dormancy to form floral organs. The process of flowering is controlled by a complex regulatory network, with at least seven flowering regulation pathways found in A. thaliana 79 . The genes associated with floral development can be divided into two classes. One class consists of genes that control the formation of inflorescence meristems and determine the direction of newly formed floral primordia. These genes influence the flowering time of plants by controlling the formation of inflorescence meristems or flower meristems, and mutations in these genes can result in earlier or later flowering mutants. The other class consists of genes that determine the formation of floral organs, and mutations in these genes can result in homeoboxes 79 . In ornamental plants, the morphology and number of floral organs have undergone substantial variations, for example, double petals, multiple sepals, and multiple pistils and stamens, developing into independent flowers during the course of long-term artificial domestication and cultivation. These variations increase the ornamental value of ornamental plants while providing excellent materials for the study of floral organ development in plants. With genomic data analysis, as an important scientific issue, some key genes related to flowering transition and flower development have been analyzed, such as those in Tarenaya hassleriana 23 , Dendrobium officinale 7 , Primula veris 28 , Dendrobium catenatum 16 , Hibiscus syriacus 41 , Rosa 4 , 5 , 12 , Chrysanthemum 18 , 19 , and Nymphaea colorata 64 .
Candidate genes for controlling anthocyanin synthesis
Flower color is one of the most vital quality traits of ornamental plants. Anthocyanin is an essential pigment for coloring flowers, and its biosynthesis is catalyzed by a series of enzymes 80 . Various anthocyanins are formed due to differences in the substituent groups at varied positions on the basic skeleton, thus leading to different plant organ colors, such as red, purple, blue-purple, and blue. Anthocyanins are flavonoid secondary metabolites in plants and the most widely distributed water-soluble pigments in nature, playing a major role in the color formation and antioxidation in plant flowers and fruits. R2R3-MYB genes are involved in anthocyanin synthesis 81 . In P. mume , 96 R2R3-MYB genes were identified and divided into 35 subfamilies. Finally, the functions of PmMYB1 and PmMYBa1 were identified by overexpression in tobacco and significantly promoted the accumulation of anthocyanins in transgenic tobacco. The flower colors of PmMYB1 -overexpressing transgenic plants were significantly deepened, and the anthocyanin contents in the corolla of transgenic plants were significantly higher than those of the control 82 . To understand the molecular basis of the blue color in water lily, delphinidin 3′-O was identified as the main blue anthocyanidin pigment, and some genes for an anthocyanidin synthase and a delphinidin-modification enzyme were screened by comparing the expression profiles between two N. colorata cultivars with white and blue petals 64 . Interestingly, after the butterfly pea UDP (uridine diphosphate)-glucose: anthocyanin 3′,5′-O-glucosyltransferase gene was introduced in chrysanthemums, blue flowers appeared 83 . In Rosa rugosa , two MYB transcription factors have been confirmed to affect flower color by regulating flavonoid biosynthesis in response to wounding and oxidation 84 . In Paeonia , a chalcone synthase ( PhCHS ) involved in flavonoid biosynthesis and two anthocyanin O-methyltransferase ( AOMT ) genes were consistent with anthocyanin accumulation in petals 85 , 86 .
Candidate genes for controlling floral scent biosynthesis
Floral scent, as one of the quality traits of ornamental plants, has great aesthetic, economic, and application value. The scent components present in petals primarily include secondary metabolites such as esters, alcohols, ketones, aldehydes, terpenes, and volatile phenols, mainly derived from terpene metabolism, phenylpropane metabolism, and the lipoxygenase pathway 87 . There are various types of scent components in different petals, thereby forming distinct scents among various flower species. In a study on the molecular mechanism responsible for the floral scent in P. mume , Zhang et al. 3 first discovered that the benzylalcohol acetyltransferase ( BEAT ) gene can directly catalyze the formation of benzyl acetate, a crucial component of the floral scent in P. mume . Moreover, based on genomic data from P. mume and P. persica , 44 unique PmBEATs were found in P. mume , far more than the 16 in apple, 14 in strawberry, and four in grape. These PmBEAT genes originated from gene duplication events during the species evolution of P. mume , and retroduplication and tandem duplication were the two dominant duplication patterns. Overexpression of the PmBEAT36 or PmBEAT37 genes increased benzyl acetate production in the petal protoplasts of P. mume , and interference in the expression of these genes slightly decreased the benzyl acetate content 88 . Zhao et al. 78 conducted a comparative transcriptome analysis of different developmental stages and tissues of flower genes associated with floral traits and preliminarily selected 12 new genes involved in floral scent formation in P. mume . Furthermore, five of the TFs ( bHLH4 , bHLH6 , bZIP4 , ERF1 , and NAC1 ) from Phalaenopsis bellina have been proven to be involved in orchid floral monoterpenes 89 . In Plumeria rubra , PrCYP79D73 is involved in floral volatile organic compounds and other nitrogen-containing volatiles 90 .
Candidate genes for controlling plant architecture
Rich and diverse plant architectures are the result of long-term evolution, natural selection, and a complex regulatory process of interaction between genetics and the environment. Diverse plant architecture traits are not only conducive to the creation of rich and diverse horticultural landscapes but are also favorable for plant adaptation to complex environments and competition and the utilization of light and nutrients. Along with the completion of whole-genome sequencing for multiple ornamental plants of the genus Prunus , the results lay an important data foundation for studying the molecular genetic mechanisms of pendulous traits 3 , 91 . According to the eight scaffolds of the P. mume genome, Zhang et al. constructed a high-density genetic map using specific-length amplified fragment sequencing (SLAF) and mapped QTLs for major traits such as plant type, flower color, petals, and leaves in P. mume . They found 10 SLAF markers that were closely linked to the pendulous traits of P. mume . Using these markers, the pendulous traits were finely mapped to a 1.14 cM region on chromosome 7, and 36 candidate genes that might be associated with the pendulous traits of P. mume were predicted 92 . Breakthroughs were also achieved in the mining and labeling of genes for weeping and dwarf traits in peach ( P. persica ) by using genome and bulked segregant analyses 93 .
Candidate genes for controlling dormancy release
Flowers of the genus Prunus , such as P. mume and P. yedoensis , are early flowering types in spring. Zhang et al. 3 explored the molecular mechanisms underpinning dormancy break and flowering in P. mume at low temperature. These researchers identified a total of six dormancy-associated MADS-box ( DAM ) genes with a tandem repeat distribution in the genome. The six DAM genes in P. mume are derived from a series of duplication events in the following order: PmDAM1 , PmDAM3 , PmDAM2 , PmDAM5 , PmDAM4 , and PmDAM6 . The molecular evolution pattern of DAM genes is unique to Prunus plants and is present in P. persica , but tandem genes have not been found in M. domestica or F. ananassa . This phenomenon could be related to the earlier flowering of Prunus plants, including P. persica , P. mume , apricot ( Armeniaca vulgaris ) and sweet cherry ( Prunus avium ), than of most other flowering species 3 . DAM genes are regulated by C-repeat-binding transcription factors (CBFs). A conserved CBF site was found 1000 bp upstream of the transcription start site of DAM4 - DAM6 in P. persica and plum ( Prunus salicina ). The latest research results show that a sense-response relationship between PmCBFs and PmDAMs is exhibited in cold-induced dormancy and is jointly regulated by six PmCBFs and PmDAM4–6 94 .
Candidate genes for controlling self-incompatibility
Self-incompatibility has always been an important research topic in the molecular genetic biology of flowers. According to different hereditary patterns of pollen incompatibility phenotypes, the regeneration disorder whereby plants reject self-pollen can be divided into sporophytic self-incompatibility and gametophytic self-incompatibility 95 . Various flowers of the Rosaceae family, including P. mume , P. yedoensis and P. persica , all exhibit gametophytic self-incompatibility, which is controlled by an S-locus with multiple alleles, including two linked genes: one is the S-RNase gene specifically expressed in pistil tissue, and the other is the S-haplotype-specific F-box gene specifically expressed in pollen 96 . In Tarenaya hassleriana , three syntenic regions containing most of the genes of the S-locus were found, and it was assumed that the single-copy ancestral region contained homologs of Pub8 , ARK3 , and B120 23 .
Candidate genes for controlling disease resistance
Disease resistance is an essential trait that attracts research attention across all flowering plants. Thus, the whole-genome analysis also focuses on the genes associated with disease resistance. The genes involved in plant disease resistance are mainly R genes, which encode proteins with extremely high structural similarities, such as leucine zippers, nucleotide-binding sites, transmembrane domains, leucine-rich repeats, and similar extracellular regions of drosophilid toll protein and mammalian toll and interleukin-1 receptor (TIR). Nucleotide-binding site leucine-rich repeat genes constitute the gene family with the widest distribution and largest number of plant R genes. In their encoded proteins, the nucleotide-binding site is present near the N-terminus, while the leucine-rich repeat exists near the C-terminus. The N-terminus of proteins encoded by different genes may also include one or more of the following two conserved structures: the coiled-coil motif and TIR motif. In the P. mume genome, 253 leucine-rich repeats receptor-like kinase (LRR-RLK) genes were identified, and most pathogenesis-related (PR) gene families were notably expanded and arranged in tandem, especially PR10 3 . In Hibiscus syriacus , resistance (R) genes account for 0.53% of its total predicted genes, which is lower than that of other plants evaluated in genomic studies (0.63 to 1.35%) 41 . The Asparagus setaceus genome included 76 R genes with nucleotide-binding sites (NBSs), and the R genes belonged to five groups: TIR-NBS, CC-NBS-LRR, NBS-LRR, NBS, and CC-NBS. NBS-LRR was the largest group, including a total of 29 genes 65 .
Candidate genes for controlling abiotic stress resistance
Adverse conditions such as low temperature, humidity, heat, drought, and saline-alkali conditions severely inhibit the growth and development of ornamental plants. These conditions can cause changes in plant physiology, biochemistry, and morphology and even lead to death. Due to this issue, cultivation facilities for ornamental plants are cumbersome and cannot be widely promoted, which considerably affects their qualities and benefits. Low temperature is an important factor that constrains the normal growth, development, and geographical distribution of plants. Stress caused by low temperature can be divided into chilling stress (>0 °C) and freezing stress (<0 °C). Plants from the tropics and subtropics are more sensitive to cold; in contrast, plants from temperate regions have evolved complex mechanisms to resist and adapt to chilling (freezing) stress, protecting the plants from injury. Cold acclimation is a responsive protection mechanism for plant adaptation and resistance to low-temperature stress, and this process is regulated by a complex network 97 . In particular, the CBF pathway is considered the most important and well-studied pathway 98 . Based on the genome data for P. mume , 30 LEA genes were identified, and heterologous expression of PmLEA increased the cold resistance of Escherichia coli and tobacco ( Nicotiana tabacum ) 99 , 100 . Furthermore, a molecular regulation model of the PmDAM and PmCBF genes in response to dormancy and dormancy release of flower buds induced by low-temperature signals was proposed based on yeast two-hybrid and bimolecular fluorescence complementation experiments 94 .
Prospects for whole-genome sequencing data for ornamental plants
The Earth BioGenome Project (EBP) is a massive project in biology that aims to sequence, catalog, and characterize the genomes of all of Earth’s eukaryotic biodiversity over a period of 10 years. For plants, the core scientific problems are to improve crop yields and other agronomically important traits, biofuel production, gene editing, and conservation of endangered species 101 . The 10,000 Plant Genome Sequencing Project (10KP) initiated by the Beijing Genomics Institute in Shenzhen (BGI-Shenzhen) is a landmark effort to catalog plant genomic variation and represents a major step in understanding the tree of life 102 . A tentative plan of the 100 Flowers Genome Sequencing Project has been put forward by the National Engineering Research Center for Floriculture in China. Many ornamentals are marked by high ploidy levels and homologous polyploids (chrysanthemum and alfalfa) or extremely large genome sizes (lily and tulip), which limit the development and utilization of genome sequencing technology in ornamental plants. Along with the development of sequencing and bioinformatics analysis technologies and the continuous emergence of various new biological technologies, genomics research on ornamental plants has developed faster and better. Although genome sequencing and assembly of flowering plants face substantial difficulties, the quality of genome assembly results is relatively high in terms of the analytical results from 69 flower species that underwent genome sequencing, and four of them have been resequenced using updated sequencing technology 5 , 11 , 37 , 50 . As far as we know, there are at least a dozen ornamental plants undergoing the process of genome quality improvement. As more ornamental plant genomes are sequenced, further bioinformatics analysis could reveal crucial basic information on the origin of species and the genes that control flower traits. The development of genomics will surely address the knowledge gaps of traditional breeding methods. The ultimate goal is to obtain the optimal type of flower variety with fixed-point improvement and the aggregation of multiple elite traits by using the most effective and rapid method.
China has 30,000 species of higher (flowering) plants, and some ornamental flowering plants reached Europe quite early 103 . Chinese people love flowers and cultivate many kinds of brilliant flowers, such as mei, peony, chrysanthemum, rose, lily, lotus, and orchid. Due to the rapid development of genome sequencing technology worldwide, large quantities of whole-genome sequencing data are in urgent need of deep mining. A long-term strategic genomics research plan should be formulated that is not limited to cultivated species but considers thorough development of the sequencing of important wild relatives of ornamental species in China and promoting the mining, protection, and utilization of important genetic resources. It is essential to put an end to the dependence on the apparent phenotype, transform investigations into genotype-dependent research and shift from single-gene studies to GWAS. Efforts should be made to vigorously promote the application of genomics in gene cloning and molecular breeding in China and to improve the breeding capacity and level of horticultural crops.
Due to their complexity and particularity, plant genomes have always been an important focus of genomics. Before the second generation of high-throughput sequencing, sequencing costs were high, and the throughput was low. For species with highly repetitive sequences, it was too difficult or too expensive for researchers to obtain the whole-genome sequences of high repeat sequence species. Many species with important economic and ornamental value have not yet been submitted to complete genome sequencing. In short, due to the particularity and diversity of ornamental plants, there are challenges and opportunities in genome research of these species. Challenge: (1) Complex genome. The term complex genome refers to a kind of genome that cannot be directly analyzed by conventional sequencing and assembly methods. It usually refers to a genome containing a high proportion of repetitive sequences, high heterozygosity, extreme GC content, and difficulty in eliminating foreign DNA contamination. (2) Autopolyploidy. Autopolyploidy is common in ornamental plants. It is usually formed by doubling two or more sets of genomes, which is of great value in genetic breeding and agricultural production. Using conventional methods, it is easy to connect incorrect allele fragments together, resulting in the wrong connection of homologous chromosomes and a large number of chimeric assemblies; thus, assembly is still difficult. (3) Megagenome. Megagenome generally refers to species with genomes larger than 10 Gb. The sequencing and analysis of these species are very involved, especially for assembly analysis, which is a major challenge. Paris japonica is an unusual plant. Scientists have found that it has the world’s largest genome, with 150 Gb, which is 50 times more than that of humans. Although the genomes of some ornamental plants have been deemed complete, the assembly quality of some species is poor, and a small number of “holes” have not yet been completed due to technical limitations, although the interest of scientists in this regard is debatable. The latest research shows that the sequences that were once considered irrelevant, or “garbage”, in the genome have their own significance. These missing sequences play a very important role, and we now have the opportunity to mine them. Third-generation sequencing technology (PacBio and Nanopore) can make up for the holes in some genomic regions that are difficult to assemble due to sequencing errors, repeat regions, heterochromatin, genomic polymorphisms, and second-generation sequencing preferences. To solve the challenge of sequencing the genomes of ornamental plants, the following new technologies can be tried with third-generation sequencing technology. (1) Pangenome. The pangenome includes the core genome and the nonessential genome. Among them, the core genome refers to the genes that exist in all individuals; the nonessential genome refers to the genes that exist only in some individuals. (2) Hi-C. The advantages of Hi-C sequencing technology are as follows: on the one hand, there is no need to construct a large number of F 1 populations, as only individuals are needed; on the other hand, the haplotype genome can be separated without parent purification, so this method is suitable for the assembly of a highly heterozygous genome that is not easy to purify.
With the development of sequencing technology, the concepts of difficult genome sequencing and assembly quality have also developed and changed. We cannot sequence everything for the sake of genome sequencing. The purpose of sequencing must be to reveal the key scientific problems of species. We should strengthen research related to transcriptomics, metabolomics, proteomics, degradomics, and phenomics. With more genomic data published, it has become a great challenge to analyze, store and share the massive amounts of genome sequencing data. A key problem is how to solve the time and cost problems faced by researchers to achieve the purpose of reducing repetitive research, improving the practicability of scientific research, mining research content, and improving the transparency of scientific research and data sharing with cross-research into other fields. Moreover, it is necessary to enhance bioinformatics education and apply bioinformatics in practice. With the continuous development of sequencing technology, we believe that the whole-genome sequencing of horticultural crops will enter a rapid development stage in the near future, leading to tremendous contributions to the world’s horticultural industry.
Initiative, A. G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408 , 796–815 (2000).
Article Google Scholar
Fan, J. Plant Genomics . (Science Press, 2019).
Zhang, Q. et al. The genome of Prunus mume . Nat. Commun. https://doi.org/10.1038/ncomms2290 (2012).
Raymond, O. et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50 , 772–777 (2018).
Article CAS PubMed PubMed Central Google Scholar
Hibrand Saint-Oyant, L. et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat Plants 4 , 473–484 (2018).
Schwartz, D. C. et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science 262 , 110–114 (1993).
Article CAS PubMed Google Scholar
Yan, L. et al. The genome of dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 8 , 922–934 (2015).
Xu, Z. et al. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. https://doi.org/10.1186/s12915-020-00795-3 (2020).
Shang, J. et al. The chromosome-level wintersweet ( Chimonanthus praecox ) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 21 , https://doi.org/10.1186/s13059-020-02088-y (2020).
Baek, S. et al. Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol. https://doi.org/10.1186/s13059-018-1497-y (2018).
Shirasawa, K. et al. Phased genome sequence of an interspecific hybrid flowering cherry, ‘Somei-Yoshino’ ( Cerasus × yedoensis ). DNA Res. 26 , 379–389 (2019).
Nakamura, N. et al. Genome structure of Rosa multiflora , a wild ancestor of cultivated roses. DNA Res. 25 , 113–121 (2018).
Cai, J. et al. The genome sequence of the orchid Phalaenopsis equestris . Nat. Genet. 47 , 65–72 (2015).
Huang, J. et al. The genome and transcriptome of Phalaenopsis yield insights into floral organ development and flowering regulation. PeerJ 4 , e2017 (2016).
Article PubMed PubMed Central CAS Google Scholar
Chao, Y. et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 16 , 2027–2041 (2018).
Zhang, G. et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. https://doi.org/10.1038/srep19029 (2016).
Zhang, G. et al. The Apostasia genome and the evolution of orchids. Nature 549 , 379–383 (2017).
Song, C. et al. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant. 11 , 1482–1491 (2018).
Hirakawa, H. et al. De novo whole-genome assembly in Chrysanthemum seticuspe , a model species of Chrysanthemums , and its application to genetic and gene discovery analysis. DNA Res. 26 , 195–203 (2019).
Badouin, H. et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546 , 148–152 (2017).
Ming, R. et al. Genome of the long-living sacred lotus ( Nelumbo nucifera Gaertn.). https://doi.org/10.1186/gb-2013-14-5-r41 (2013).
Sierro, N. et al. Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis . Genome Biol. 14 , R60 (2013).
Cheng, S. et al. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25 , 2813–2830 (2013).
Wang, Y. et al. The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 76 , 557–567 (2013).
Hellsten, U. et al. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc. Natl Acad. Sci. USA 110 , 19478–19482 (2013).
Yagi, M. et al. Sequence analysis of the genome of carnation ( Dianthus caryophyllus L.). DNA Res. 21 , 231–241 (2014).
Sunil, M. et al. The draft genome and transcriptome of Amaranthus hypochondriacus : a C4 dicot producing high-lysine edible pseudo-cereal. DNA Res. 21 , 585–602 (2014).
Nowak, M. D. et al. The draft genome of Primula veris yields insights into the molecular basis of heterostyly. Genome Biol. https://doi.org/10.1186/s13059-014-0567-z (2015).
Kellner, F. et al. Genome-guided investigation of plant natural product biosynthesis. Plant J. 82 , 680–692 (2015).
Xiao, L. et al. The resurrection genome of Boea hygrometrica : A blueprint for survival of dehydration. Proc. Natl Acad. Sci. USA 112 , 5833–5837 (2015).
Byrne, S. L. et al. A synteny-based draft genome sequence of the forage grass Lolium perenne . Plant J. 84 , 816–826 (2015).
De Vega, J. J. et al. Red clover ( Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. https://doi.org/10.1038/srep17394 (2015).
Lu, M., An, H. & Li, L. Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii tratt and leaf ascorbate metabolism genes. PLoS ONE 11 , e147530 (2016).
Google Scholar
Tanaka, H. et al. Sequencing and comparative analyses of the genomes of zoysiagrasses. DNA Res. 23 , 171–180 (2016).
Bombarely, A. et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida . Nat. Plants https://doi.org/10.1038/nplants.2016.74 (2016).
Butts, C. T., Bierma, J. C. & Martin, R. W. Novel proteases from the genome of the carnivorous plant Drosera capensis : Structural prediction and comparative analysis. Proteins 84 , 1517–1533 (2016).
Clouse, J. W. et al. The Amaranth genome: Genome, transcriptome, and physical map assembly. Plant Genome https://doi.org/10.3835/plantgenome2015.07.0062 (2016).
Hirakawa, H. et al. Draft genome sequence of subterranean clover, a reference for genus Trifolium . Sci. Rep. https://doi.org/10.1038/srep30358 (2016).
Hoshino, A. et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil . Nat. Commun. https://doi.org/10.1038/ncomms13295 (2016).
Guan, R. et al. Draft genome of the living fossil Ginkgo biloba . Gigascience https://doi.org/10.1186/s13742-016-0154-1 (2016).
Kim, Y. et al. Genome analysis of Hibiscus syriacus provides insights of polyploidization and indeterminate flowering in woody plants. DNA Res . https://doi.org/10.1093/dnares/dsw049 (2016).
Sollars, E. S. et al. Genome sequence and genetic diversity of European ash trees. Nature 541 , 212–216 (2017).
Fu, Y. et al. Draft genome sequence of the Tibetan medicinal herb Rhodiola crenulata . Gigascience https://doi.org/10.1093/gigascience/gix033 (2017).
Zhao, D. et al. De novo genome assembly of Camptotheca acuminata , a natural source of the anti-cancer compound camptothecin. Gigascience 6 , 1–7 (2017).
Zhang, L. et al. The draft genome assembly of Rhododendron delavayi Franch. var. delavayi. Gigascience https://doi.org/10.1093/gigascience/gix076 (2017).
Copetti, D. et al. Extensive gene tree discordance and hemiplasy shaped the genomes of North American columnar cacti. Proc. Natl Acad. Sci. USA 114 , 12003–12008 (2017).
Silva-Junior, O. B., Grattapaglia, D., Novaes, E. & Collevatti, R. G. Genome assembly of the Pink Ipê (Handroanthus impetiginosus, Bignoniaceae), a highly valued, ecologically keystone Neotropical timber forest tree. Gigascience https://doi.org/10.1093/gigascience/gix125 (2018).
Yang, X. et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. https://doi.org/10.1038/s41467-017-01491-7 (2017).
Hori, K. et al. Mining of the uncharacterized cytochrome P450 genes involved in alkaloid biosynthesis in california poppy using a draft genome sequence. Plant Cell Physiol. 59 , 222–233 (2018).
Gui, S. et al. Improving Nelumbo nucifera genome assemblies using high‐resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 94 , 721–734 (2018).
Gao, Y. et al. De novo genome assembly of the red silk cotton tree ( Bombax ceiba ). Gigascience https://doi.org/10.1093/gigascience/giy051 (2018).
Dong, A. et al. High-quality assembly of the reference genome for scarlet sage, Salvia splendens , an economically important ornamental plant. Gigascience https://doi.org/10.1093/gigascience/giy068 (2018).
Griesmann, M. et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361 , https://doi.org/10.1126/science.aat1743 (2018).
Malli, R. P. N., Adal, A. M., Sarker, L. S., Liang, P. & Mahmoud, S. S. De novo sequencing of the Lavandula angustifolia genome reveals highly duplicated and optimized features for essential oil production. Planta 249 , 251–256 (2019).
Ye, G. et al. De novo genome assembly of the stress tolerant forest species Casuarina equisetifolia provides insight into secondary growth. Plant J. 97 , 779–794 (2019).
Yang, X. et al. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans . Hortic. Res. https://doi.org/10.1038/s41438-018-0108-0 (2018).
Chen, J. et al. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 5 , 18–25 (2019).
Cocker, J. M. et al. Primula vulgaris (primrose) genome assembly, annotation and gene expression, with comparative genomics on the heterostyly supergene. Sci. Rep. https://doi.org/10.1038/s41598-018-36304-4 (2018).
Li, M. et al. Genome structure and evolution of Antirrhinum majus L. Nat. Plants 5 , 174–183 (2019).
Wai, C. M. et al. Time of day and network reprogramming during drought induced CAM photosynthesis in Sedum album . PLoS Genet. 15 , e1008209 (2019).
Soza, V. L. et al. The Rhododendron genome and chromosomal organization provide insight into shared whole-genome duplications across the heath family (Ericaceae). Genome Biol. Evol. 11 , 3353–3371 (2019).
Yamashiro, T., Shiraishi, A., Satake, H. & Nakayama, K. Draft genome of Tanacetum cinerariifolium , the natural source of mosquito coil. Sci. Rep. https://doi.org/10.1038/s41598-019-54815-6 (2019).
Lv, S. et al. Draft genome of the famous ornamental plant Paeonia suffruticosa . Ecol. Evol. 10 , 4518–4530 (2020).
Article PubMed PubMed Central Google Scholar
Zhang, L. et al. The water lily genome and the early evolution of flowering plants. Nature. 577 , 79–84 (2020).
Li, S. et al. Chromosome-level genome assembly, annotation and evolutionary analysis of the ornamental plant Asparagus setaceus . Hortic. Res. https://doi.org/10.1038/s41438-020-0271-y (2020).
Palfalvi, G. et al. Genomes of the venus flytrap and close relatives unveil the roots of plant carnivory. Curr. Biol. 30 , 2312–2320 (2020).
Lv, Q. et al. The Chimonanthus salicifolius genome provides insight into magnoliid evolution and flavonoid biosynthesis. Plant J. 103 , 1910–1923 (2020).
Li, L., Cushman, S. A., He, Y. & Li, Y. Genome sequencing and population genomics modeling provide insights into the local adaptation of weeping forsythia. Hortic. Res. https://doi.org/10.1038/s41438-020-00352-7 (2020).
Yi, X. et al. The genome of Chinese flowering cherry ( Cerasus serrulata ) provides new insights into Cerasus species. Hortic. Res. https://doi.org/10.1038/s41438-020-00382-1 (2020).
Yang, F. S. et al. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat. Commun. 11 , 5269 (2020).
Zhou, Z. et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33 , 408–414 (2015).
Varshney, R. K. et al. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 49 , 1082–1088 (2017).
Hübner, S. et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 5 , 54–62 (2019).
Article PubMed CAS Google Scholar
Zhang, Q. et al. The genetic architecture of floral traits in the woody plant Prunus mume . Nat. Commun. https://doi.org/10.1038/s41467-018-04093-z (2018).
Huang, L. et al. Whole genome re-sequencing reveals evolutionary patterns of sacred lotus ( Nelumbo nucifera ). J. Integr. Plant Biol. 60 , 2–15 (2018).
Article PubMed Google Scholar
Sasaki, R. et al. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157 , 485–497 (2011).
Jimenez, S., Lawtonrauh, A., Reighard, G. L., Abbott, A. G. & Bielenberg, D. G. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 9 , 81 (2009).
Zhao, K. et al. Comparative transcriptome reveals benzenoid biosynthesis regulation as inducer of floral scent in the woody plant Prunus mume . Front. Plant Sci. https://doi.org/10.3389/fpls.2017.00319 (2017).
Fornara, F., De Montaigu, A. & Coupland, G. SnapShot: control of flowering in Arabidopsis . Cell https://doi.org/10.1016/j.cell.2010.04.024 (2010).
Tanaka, Y. & Ohmiya, A. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotech. 19 , 190–197 (2008).
Xu, W., Dubos, C. & Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20 , 176–185 (2015).
Zhang, Q. et al. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell Tiss. Org. 131 , 417–429 (2017).
Article CAS Google Scholar
Noda, N. et al. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci. Adv. 3 , e1602785 (2017).
Shen, Y. et al. RrMYB5 and RrMYB10 regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa . Plant Biotechnol. J. 17 , 2078–2095 (2019).
Du, H. et al. Methylation mediated by an anthocyanin, O -methyltransferase, is involved in purple flower coloration in. Paeonia. J. Exp. Bot. 66 , 6563–6577 (2015).
Gu, Z. et al. Chalcone synthase is ubiquitinated and degraded via interactions with a RING-H2 protein in petals of Paeonia ‘He Xie’. J. Exp. Bot. 70 , 4749–4762 (2019).
Dudareva, N. & Pichersky, E. Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 122 , 627–633 (2000).
Bao, F. et al. Expansion of PmBEAT genes in the Prunus mume genome induces characteristic floral scent production. Hortic. Res. 6 , 24 (2019).
Chuang, Y. C., Hung, Y. C., Tsai, W. C., Chen, W. H. & Chen, H. H. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids . J. Exp. Bot. 69 , 4363–4377 (2018).
Dhandapani, S., Jin, J., Sridhar, V., Chua, N. & Jang, I. CYP79D73 participates in biosynthesis of floral scent compound 2-Phenylethanol in Plumeria rubra . Plant Physiol. 180 , 171–184 (2019).
Verde, I. et al. The high-quality draft genome of peach ( Prunus persica ) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45 , 487–494 (2013).
Zhang, J. et al. High-density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant ( Prunus mume Sieb. et Zucc). DNA Res. 22 , 183–191 (2015).
Hollender, C. A. et al. Loss of a highly conserved sterile alpha motif domain gene ( WEEP ) results in pendulous branch growth in peach trees. Proc. Natl Acad. Sci. USA 115 , 201704515 (2018).
Zhao, K. et al. Crosstalk of PmCBFs and PmDAMs Based on the changes of phytohormones under seasonal cold stress in the stem of Prunus mume . Int. J. Mol. Sci. 19 , 15 (2018).
Article PubMed Central CAS Google Scholar
Mcclure, B. & Franklintong, V. E. Gametophytic self-incompatibility: understanding the cellular mechanisms involved in “self” pollen tube inhibition. Planta 224 , 233–245 (2006).
Donia, A., Ghada, B., Hend, B. T., Sana, B. M. & Amel, S. H. Identification, evolutionary patterns and intragenic recombination of the gametophytic self incompatibility pollen gene ( SFB ) in tunisian Prunus species (Rosaceae). Plant Mol. Biol. Rep. 34 , 339–352 (2016).
Shi, Y., Ding, Y. & Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23 , 623–637 (2018).
Liu, J., Shi, Y. & Yang, S. Insights into the regulation of C‐repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60 , 780–795 (2018).
Bao, F. et al. Overexpression of Prunus mume dehydrin genes in tobacco enhances tolerance to cold and drought. Front. Plant Sci. 8 , 151 (2017).
Du, D. et al. Genome-wide identification and analysis of late embryogenesis abundant ( LEA ) genes in Prunus mume . Mol. Biol. Rep. 40 , 1937–1946 (2013).
Lewin, H. A. et al. Earth BioGenome Project: sequencing life for the future of life. Proc. Natl Acad. Sci. USA 115 , 4325–4333 (2018).
Twyford, A. D. The road to 10,000 plant genomes. Nat. Plants 4 , 312–313 (2018).
Wilson, E. H. China, Mother of Gardens . (The Stratford Company, 1929).
Download references
Acknowledgements
The research was supported by the National Natural Science Foundation of China (No. 31800595 and 31471906), the National Key Research and Development Program of China (2018YFD1000401), and the Special Fund for Beijing Common Construction Project.
Author information
Authors and affiliations.
Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Key Laboratory of Ornamental Plants Germplasm Innovation & Molecular Breeding, National Engineering Research Center for Floriculture, Beijing Laboratory of Urban and Rural Ecological Environment, Engineering Research Center of Landscape Environment of Ministry of Education, Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants of Ministry of Education, School of Landscape Architecture, Beijing Forestry University, Beijing, 100083, China
Tangchun Zheng, Ping Li, Lulu Li & Qixiang Zhang
You can also search for this author in PubMed Google Scholar
Contributions
T.Z. conceived and drafted the manuscript. T.Z., P.L., and L.L. analyzed the data. Q.Z. contributed to the conception of the study and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Qixiang Zhang .
Ethics declarations
Conflict of interest.
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Zheng, T., Li, P., Li, L. et al. Research advances in and prospects of ornamental plant genomics. Hortic Res 8 , 65 (2021). https://doi.org/10.1038/s41438-021-00499-x
Download citation
Received : 08 September 2020
Revised : 04 January 2021
Accepted : 11 January 2021
Published : 01 April 2021
DOI : https://doi.org/10.1038/s41438-021-00499-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Breeding of ornamental orchids with focus on phalaenopsis: current approaches, tools, and challenges for this century.
- Carla Midori Iiyama
- Joe Abdul Vilcherrez-Atoche
- Jean Carlos Cardoso
Heredity (2024)
QTL Mapping and Genetic Map for the Ornamental Sunflower in China
- Junjian Shan
Plant Molecular Biology Reporter (2024)
Single-molecule long-read sequencing analysis improves genome annotation and sheds new light on the transcripts and splice isoforms of Zoysia japonica
BMC Plant Biology (2022)
Characterization and expression analysis of WRKY genes during leaf and corolla senescence of Petunia hybrida plants
- Francisco H. Astigueta
- Amilcar H. Baigorria
- Santiago A. Trupkin
Physiology and Molecular Biology of Plants (2022)
CRISPR/Cas9 System: A Potential Tool for Genetic Improvement in Floricultural Crops
- Ujjwal Sirohi
- Mukesh Kumar
- Manoj Kumar Yadav
Molecular Biotechnology (2022)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Int J Environ Res Public Health

Effects of Indoor Plants on Human Functions: A Systematic Review with Meta-Analyses
Associated data.
Data are available upon reasonable request.
The influences of indoor plants on people have been examined by only three systematic reviews and no meta-analyses. The objective of this study was therefore to investigate the effects of indoor plants on individuals’ physiological, cognitive, health-related, and behavioral functions by conducting a systematic review with meta-analyses to fill the research gap. The eligibility criteria of this study were (1) any type of participants, (2) any type of indoor plants, (3) comparators without any plants or with other elements, (4) any type of objective human function outcomes, (5) any type of study design, and (6) publications in either English or Chinese. Records were extracted from the Web of Science (1990–), Scopus (1970–), WANFANG DATA (1980–), and Taiwan Periodical Literature (1970–). Therefore, at least two databases were searched in English and in Chinese—two of the most common languages in the world. The last search date of all four databases was on 18 February 2021. We used a quality appraisal system to evaluate the included records. A total of 42 records was included for the systematic review, which concluded that indoor plants affect participants’ functions positively, particularly those of relaxed physiology and enhanced cognition. Separate meta-analyses were then conducted for the effects of the absence or presence of indoor plants on human functions. The meta-analyses comprised only 16 records. The evidence synthesis showed that indoor plants can significantly benefit participants’ diastolic blood pressure (−2.526, 95% CI −4.142, −0.909) and academic achievement (0.534, 95% CI 0.167, 0.901), whereas indoor plants also affected participants’ electroencephalography (EEG) α and β waves, attention, and response time, though not significantly. The major limitations of this study were that we did not include the grey literature and used only two or three records for the meta-analysis of each function. In brief, to achieve the healthy city for people’s health and effective functioning, not only are green spaces needed in cities, but also plants are needed in buildings.
1. Introduction
Throughout history, humans have valued the health benefits of contact with nature [ 1 ]. Theoretically, an evolutionary perspective suggests that evolutionary processes enable humans to respond adaptively and positively to nature [ 2 ], whereas a cultural perspective contends that culture affects people’s relations with the natural environment [ 3 ]. In line with the evolutionary perspective, the concept of biophilia claims that humans are born with emotional connections with nature and/or other living organisms [ 4 ]. This emotional predisposition is deeply embedded in the biological nature of humans and does not disappear even after people leave the natural environment to live a modern urban life [ 5 ]. Moreover, the Stress Reduction Theory (SRT; [ 3 ]) emphasizes that stress is “the process by which an individual responds psychologically, physiologically, and often with behaviors, to a situation that challenges or threatens well-being” ([ 6 ], p. 202), and the natural environment is helpful for recovery from stress, whereas the Attention Restoration Theory (ART; [ 7 ]) emphasizes that the natural environment is beneficial to the restoration of directed attention for people’s effective functioning. Empirically, an increasing number of studies on human interaction with nature have demonstrated that contact with nature is favorable to human emotions, physiological functioning, attention restoration, behavior, and health [ 8 , 9 , 10 ]. Scholars have also conducted systematic reviews (e.g., [ 11 , 12 , 13 , 14 , 15 ] and meta-analyses on related topics (e.g., [ 16 , 17 , 18 , 19 , 20 , 21 ]). In total, by 2022, more than 60 reviews and meta-analyses regarding nature and health and well-being had been conducted (cf. [ 22 ]).
Despite long interests in the theoretical and empirical value of nature to humans, at present, 55% of the world population lives in cities, and the urban population worldwide is expected to increase by 68% by 2050 [ 23 ]. For this reason, the World Health Organization Regional Office for Europe in 2016 [ 24 ] published a report titled “Urban Green Spaces and Health—A Review of Evidence” to address the importance of nature and green spaces for urban living. The World Health Organization also advocates the healthy city, defined as “one that continually creates and improves its physical and social environments and expands the community resources that enable people to mutually support each other in performing all the functions of life and developing to their maximum potential” [ 25 ]. Individuals in contemporary society nevertheless spend most of their time indoors [ 26 ], with urban dwellers spending more than 80% of their life indoors [ 27 , 28 , 29 ]. Moreover, urbanities often do not have ready access to nature [ 30 ]. Consequently, urbanites have few opportunities to maintain contact with nature. Although nature includes many elements, plants are the most representative symbol of nature [ 31 , 32 ]. Similarly, “green space” refers to open, underdeveloped, naturally planted land [ 33 ], land with grass or trees, or other vegetation region [ 34 ]. Studies of indoor nature also tend to focus on plants [ 35 , 36 ]. The exploration of the physical and psychological benefits of indoor plants on people, therefore, merits more attention [ 37 , 38 ]. Interior environments and indoor plants could be important elements of the healthy city. The Rural Development Administration of South Korea suggests placing one small potted plant and one large potted plant per 6 m 2 floor area in a room to improve the indoor quality [ 39 ]. Systematic reviews and/or meta-analyses of the effects of indoor plants on people, however, are far less common than those on natural environments and/or green spaces.
There are only three narrative reviews on the influences of indoor plants related to people. Bringslimark et al. [ 31 ] reviewed 21 articles of the experimental research focusing on the benefits of indoor plants on people, which identified benefits such as stress reduction and pain tolerance enhancement. This study was a great stepping stone for later research, particularly with respect to experimental design, measurement, analysis, and reporting. Given that this narrative review was published more than a decade ago, updates are necessary, as is a further distinction of the benefits identified as either self-reported perceptions or objectively measured outcomes using devices or tasks. Deng and Deng [ 40 ] reviewed the importance of indoor plants to human health with respect to photosynthesis, transpiration, psychological effects, and air purification, indicating the influence of indoor plants on task performance, health, and stress. Moya et al. [ 41 ] reviewed 104 articles published in specific journals between 1984 and 2017 on the influence of vegetation on indoor environmental quality, finding that indoor plants improved people’s comfort, satisfaction, and happiness but presented no strong evidence of improvements in performance and productivity. The inconsistent findings on the influences of indoor plants on participant’s performance [ 40 , 41 ] await further clarification. Further, these three narrative reviews did not follow the rigorous Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA; [ 42 ]), which may have resulted in subjectivity and a lack of transparency and comprehensiveness [ 43 ]. They also did not cover studies published in widely used languages, such as Chinese.
Recently, three systematic reviews were conducted to address the gaps left by previous narrative reviews of research on indoor plants and human responses. One followed the Centre for Reviews and Dissemination (CRD) guidelines [ 44 ], and two followed the PRISMA guidelines. The first systematic review [ 45 ] following the PRISMA covered studies published in English and Chinese—two of the most common languages in the world—focused on self-reported perceptions and included 50 empirical studies, which concluded that the primary beneficial effects of indoor plants were an increase in positive emotions and a reduction in negative feelings, while secondary benefits included a reduction in physical discomfort. The second systematic review [ 36 ] following the CRD covered 26 studies of the health and well-being impacts of indoor nature (actual and simulated plants and aquariums) on the elderly and concluded that higher-quality studies showed that indoor gardening programs were helpful for cognition, psychological well-being, social outcomes, and life satisfaction. The third review [ 35 ] following the PRISMA covered 37 studies published in English and Dutch on the influences of indoor and outdoor nature on adolescents, which found associations between outdoor campus green space and enhanced quality of life and perceived restoration. The common findings of the two systematic reviews are that indoor plants benefit psychological well-being [ 36 , 45 ]. Self-reported psychological responses, however, may be different from actual human functions (cf. [ 46 ]). Moreover, Yeo et al. [ 36 ] researched only older adults and did not specifically focus on indoor plants. Van den Bogerd et al. [ 35 ] researched only adolescents and did not specifically focus on indoor plants. Furthermore, none of these three reviews conducted meta-analyses to provide quantitatively synthesized evidence of the effects of indoor plants on humans. This may be because of the heterogeneity of the outcomes.
Given the above-mentioned factors, the purpose of the present study was to perform a systematic review with meta-analyses of Chinese and English empirical quantitative research on the influences of indoor plants on human functions, in order to address the current research gap of the lack of meta-analyses on this subject and to respond to the fulfillment of the daily functions of urbanites as advocated by the promotion of the healthy city. Specifically, the objective of this study was to examine if the presence of indoor plants of any type serving as an intervention has any objectively measured effects, such as using devices, tasks, examinations, or performance records, on human functions. We reviewed all research with any study design that assessed all human functions exposed to indoor plants against those exposed to no indoor plants or other elements. Accordingly, the review question was whether indoor plants have any effects on human functions. The present systematic review may provide more comprehensive information associated with the aforementioned effects and serve as a reference for future research. This study identifies what empirical and quantitative studies of human functions have been performed in relation to indoor plants, particularly regarding research validity (cf. [ 31 ]), such as plant quantity measurements (construct validity: number, size, volume percentage, and green coverage ratio), potential effect modifiers (confounder: exposure duration, distance to plants, room climate, and room size), funding (conflict of interest), and what and how further research could be performed. The meta-analyses further examined the overall results of studies exploring the effects of indoor plants on human functions, rather than reviewing studies individually. Systemic reviews and meta-analyses are effective in providing optimal synthesis evidence, and the constantly updated data may serve as a basis for policy making [ 43 ], such as regarding the healthy city or effective human functions [ 25 ]. The systematic review and meta-analyses conducted in this study are the first to provide a synthesis of the quantitative evidence regarding the specific effects of indoor plants on human functions.
This study followed the PRISMA guidelines, particularly their specific checklist items and item orders, although PRISMA focuses on evaluating interventions in the field of medical care [ 42 ]. The conduct of the review involved no significant deviations from the protocol, except that we searched two more databases than the protocol.
2.1. Eligibility Criteria
The eligibility criteria for inclusion of a study in this research were as follows: (1) participants of any type were recruited; (2) no criteria were set for the type of indoor plant to be used in interventions; (3) the comparator was participants in an indoor environment without any plants or with other elements; (4) the outcome included any type of objectively measured human function, such as use of devices, performance tasks, examinations, or records, rather than self-reports; (5) all types of study design were included; and (6) the language was either English or Chinese. Because Chinese and English are the most commonly used languages in the world, studies written in these two languages were selected to decrease the risk of language bias [ 43 ]. Other languages were not included because of limited resources.
2.2. Information Sources
The information sources included four electronic databases, of which the Core Collections hosted by Web of Science (1988–) and Scopus (1970–) are English-language databases, while the Journal Collections hosted by WANFANG DATA (1980–) and Taiwan Periodical Literature (1970–) are Chinese-language databases. At least two databases, therefore, were searched for each of the two languages. The final search on WANFANG DATA was performed on 14 August 2019, while that on the Web of Science was performed on 11 November 2019. The search on Taiwan Periodical Literature was performed on 21 October 2020, and the search on Scopus was performed on 13 November 2020. We searched WANFANG DATA and Web of Science in the first round and Taiwan Periodical Literature and Scopus in the second. About one year thus elapsed between the searches of the two rounds. The follow-up searches of all four databases were completed on 18 February 2021. The coverage cutoff date was 31 December 2020. We decided that the coverage ended at the end of 2020 rather than in the middle of the year, so later updated searches could continue at the start of 2021. Moreover, two supplementary approaches to identifying studies were applied: one was that the related studies were identified by reference searches of the included studies, while The other was contact with the authors of the included studies to seek missing information, particularly regarding the results.
2.3. Search
The search terms included the following: “indoor”, “interior”, “architecture”, “building”, “plant”, “vegetation”, “greening”, “greenery”, “green”, “greenness”, “perception”, “psychology”, “emotion”, “physiology”, “cognition”, “restoration”, “behavior”, “health”, and “performance”, as found in previous studies [ 8 , 45 , 47 ] and in peer reviewers’ suggestions. In the Boolean search, only “AND” was adopted as the operator, as in (1) indoor “AND” plant “AND” perception, or (2) architecture “AND” greening “AND” psychology. Except for the eligibility criteria and the search coverage, we did not have any restrictions such as topics, keywords, or dates. The full search strings applicable to all four databases are listed in Supplementary Material Table S1 .
2.4. Study Selection
The present systematic review included only quantitative empirical studies published in journals, primarily because of their relatively easy accessibility. Technical reports, proceedings, books, and unpublished theses or dissertations (i.e., grey literature) therefore were not considered. Empirical studies using plants in a room or building as the intervention, irrespective of how many plants, what sizes, what types, foliage or floral, actual or virtual, duration of presence, or distance from the participants, were included in accordance with the eligibility criteria. “Empirical research” refers to the analysis of real data, and quantitative research uses computation, mathematics, and statistics to explore the target phenomenon [ 48 ]. Regarding the causal relationship between variables in quantitative research, randomized controlled experiments, in which participants are randomly assigned to experimental and control groups (also referred to as “RCT” in clinical professions), provide higher-quality results than nonrandomized controlled quasi-experiments, in which participants are not randomly assigned to experimental and control groups (also referred to as “non-RCT”), and quasi-experiments outperform surveys [ 49 ]. Field experiments conducted in real-world environments, however, exhibit more favorable ecological validity than laboratory experiments [ 50 ]. If surveys were used to collect objective outcomes such as health indicators, they were also included.

2.5. Data Collection Process
L.-W.R. performed searches with the abovementioned terms in the databases and reviewed study titles and abstracts that met the eligibility criteria. L.-S.L. independently performed searches with the same terms in the same databases and reviewed study titles and abstracts that met the eligibility criteria. As a result, L.-W.R. and L.-S.L. had an agreement rate of 99.9% on both Web of Science and WANFANG DATA, respectively, and L.-S.L. and K.-T.H. also had an agreement rate of 99.9% on Taiwan Periodical Literature. In cases where the title and abstract were insufficient to determine the study’s eligibility, L.-W.R. or L.-S.L. proceeded to read the full text. All the studies of each included paper were reviewed. Then, K.-T.H. reviewed the extracted full-text studies that met the eligibility criteria. K.-T.H. and L.-W.R. conducted data extraction and quality appraisal. Initial disagreement regarding a study’s opinion was resolved by discussion between the two reviewers.
2.6. Data Items
The following 14 data items were extracted from the reviewed records: sources, participants, interventions, comparator, exposure duration, distance to plants, room climate, room size, study design, functions, function categories, outcomes, funding, and languages.
2.7. Risk of Bias in Individual Studies
The included studies were analyzed in accordance with the quality appraisal system proposed by Ohly et al. [ 19 ]. This appraisal system comprises 19 appraisal items, including quality indicators from the CRD [ 44 ], critical appraisal checklists from the Critical Appraisal Skills Program [ 51 ], and quality assessment tool for quantitative studies from the Effective Public Health Practice Project [ 52 ]. We adopted the quality appraisal system because it was more comprehensive and current than other appraisal systems. This appraisal system, which has an option of criterion inapplicable to this study design, was applied to both RCTs and nonrandomized studies [ 19 , 20 ].
2.8. Summary Measures
The summary measures of this study included data measured in empirical research using devices, tasks, academic achievement scoring, and actual health indicators. These data were reviewed to examine the influences of indoor plants on participants’ functions.
2.9. Planned Methods of Analysis
Where sufficient studies (at least two studies) using comparable outcome measures allowed us to conduct meta-analyses, the present meta-analyses reported the means and standard deviations (SDs) of each function category. Comprehensive Meta-Analysis version 3 (Biostat, Englewood, NJ, USA) was applied to conduct Cochran’s Q tests and draw forest plots as well as to analyze pooled effect sizes, sensitivities, and publication biases. Because the measurement of functions in similar categories varied between the records, the measurement outcomes of these categories were processed using the standardized mean difference (SMD), thereby providing an indicator enabling the comparison and synthesis of function outcomes in these categories (cf. [ 43 ]). For records using the same methods to measure function in the same categories, outcomes were, in general, directly compared and synthesized to determine the mean differences (MDs). For studies with more than one experimental group [ 53 ], each experimental group and control group was separately analyzed [ 43 ]. Specifically, Cochran’s Q test was performed to examine whether the classification outcomes of function in each category were heterogeneous or homogenous. Additionally, forest plots were adopted to present the relative importance and research outcome directions among the studies visually. Subsequently, the pooled effect size was computed using fixed-effect or random-effect models.
2.10. Risk of Bias across Studies
The meta-analyses used funnel plots and Egger’s regressions to identify potential publication bias on the basis of the research results of each function category.
2.11. Additional Analyses
The meta-analyses included the sensitivity analysis of the results of the records for each function category, which examined if the pooled effect sizes changed notably when any of the records was removed—an indication of the stability of the results.
3.1. Study Selection
The abovementioned terms were used to search the four databases separately. The search yielded 30,887 records from the Web of Science, 4323 from WANFANG DATA, 30,203 from Scopus, and 4105 from Taiwan Periodical Literature. Repeated records were excluded. Moreover, 11 records were identified after searching the references of the searched papers (which were considered as other sources), resulting in 31,728 journal articles in total. Papers with titles and abstracts meeting the eligibility criteria were identified, resulting in 63 preliminarily qualified papers. The full texts of these 63 papers were extracted for further scrutiny, and 21 studies that failed to meet the criteria were excluded. As a result, 42 qualifying records were included ( Figure 1 ). The major reasons for excluding records were that the research was not empirical and quantitative, human functions were not objectively measured, and plant functions were measured ( Supplementary Material Table S2 ).

Flow chart of the screening process.
3.2. Study Characteristics
Among the 42 journal articles included in the systematic review, 5 (11.9%) and 37 (88.1%) were written in Chinese and English, respectively. The earliest paper was published in 1996, and the latest in 2020 (the terminus of the search coverage). The 25 year coverage period was divided into 5 year intervals to analyze the number of publications during each interval. The number of published papers increased relatively steadily ( Table 1 ).
Statistics of published journal articles in Chinese and English during consecutive 5 year periods.
| Publication Year | Publication Language | Total | ||||
|---|---|---|---|---|---|---|
| Chinese | English | |||||
| Number of Papers | Percentage (%) | Number of Papers | Percentage (%) | Number of Papers | Percentage (%) | |
| 1996–2000 | 1 | 20 | 3 | 8.1 | 4 | 9.5 |
| 2001–2005 | 0 | 0 | 6 | 16.2 | 6 | 14.3 |
| 2006–2010 | 0 | 0 | 7 | 18.9 | 7 | 16.7 |
| 2011–2015 | 1 | 20 | 9 | 24.3 | 10 | 23.8 |
| 2016–2020 | 3 | 60 | 12 | 32.4 | 15 | 35.7 |
| Total | 5 | 100.0 | 37 | 100.0 | 42 | 100.0 |
In terms of geographical distribution, most of the studies were from China (10; 23.8%), followed by the United States (8; 19.0%), Japan (6; 14.3%), South Korea (5; 11.9%), and Taiwan (4; 9.5%). Asia was thus the leading continent, followed by America, Europe, and Africa. Most studies were from the Global North, followed by the Global South and the Equatorial region ( Table 2 ). Detailed statistics could not be compiled because not every record provided the participants’ socioeconomic backgrounds. The majority of the participants, however, were college students. Only six studies recruited office workers [ 54 , 55 , 56 , 57 , 58 , 59 ], five studies recruited patients [ 60 , 61 , 62 , 63 , 64 ], two studies recruited junior high school students [ 65 , 66 ], and one study recruited high school students as participants [ 67 ] ( Table 2 ).
Statistics of geographical distribution of the included studies.
| Participant Location | Number of Records | Percentage (%) |
|---|---|---|
| China (Asia, Global North) | 10 | 23.8 |
| United States (America, Global North) | 8 | 19.0 |
| Japan (Asia, Global North) | 6 | 14.3 |
| South Korea (Asia, Global North) | 5 | 11.9 |
| Taiwan (Asia, Global North) | 4 | 9.5 |
| Norway (Europe, Global North) | 3 | 7.1 |
| United Kingdom (Europe, Global North) | 1 | 2.4 |
| Sweden (Europe, Global North) | 1 | 2.4 |
| Pakistan (Asia, Global North) | 1 | 2.4 |
| Egypt (Africa, Global North) | 1 | 2.4 |
| South Africans (Africa, Global South) | 1 | 2.4 |
| Indonesia (Asia, Equatorial) | 1 | 2.4 |
| Total | 42 | 100.0 |
The records generally did not focus on only one measure of human functions. The reviewers found that they examined 52 functions. Most of the records were related to physiology (27; 51.9%), followed by cognition (15; 28.8%), health, (seven; 13.5%), and behavior (three; 5.8%). Concerning study design, most of the records adopted experimental methods (26; 61.9%). These were followed by those conducting field experiments (seven; 16.7%), field quasi-experiments (five; 11.9%), and surveys (four; 9.5%). Among the 42 records, 20 reported the specific number of indoor potted plants as the intervention; the highest number of potted plants was 34 [ 66 ], and the lowest number of potted plants was one [ 56 , 59 , 60 , 68 , 69 , 70 , 71 ]. Three papers reported the green coverage ratio, with the highest at 10% and the lowest at 3% [ 57 ]. Two papers also indicated the volume of indoor plants as a percentage of the total experimental space, where the largest volume was 17.9% [ 53 ] and the smallest was 5% [ 67 ] ( Table 3 ). In addition, three records used photographs or slides as surrogates for indoor plants [ 72 , 73 , 74 ], and one paper employed virtual-reality plants [ 75 ] ( Table 4 ).
Summary of the study characteristics of the records.
| Source | Participant | Interventions | Comparator | Exposure Duration | Distance to Plants | Room Size | Room Climate | Study Design | Functions | Function Category | Funding | Publication Language |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [ ] | 96 US adults (48 males and 48 females, 80 of whom were college students), age: 18 to 46 | Presence or absence of 17 potted plants in a computer lab | Control | 13.5 × 7.3 × 2.6 m | 27 °C, 38% RH, 420 lux | Field experiment (RCT) | SBP, reaction time | Physiology, cognition | English | |||
| [ ] | 81 US adults | 10 potted plants (accounting 7.16% of the space), 22 potted plants (accounting 17.88% of the space), or no plants in an office | Control | 15–20 min | 12.08 m , 31.3 m | Field experiment (RCT) | A sorting task, a productivity task | Cognition | English | |||
| [ ] | 814 Chinese participants (347 males and 467 females), ethnicity: Asian | A building with or without indoor greening | Survey (non-RCT) | Neurobehavioral Functioning Evaluation System Testing | Cognition | Sciences and Technology Commission of Shanghai | Chinese | |||||
| [ ] | 198 US adults (71 males and 127 females), 176 of whom were college students | 5 potted plants, nonplant objects, no plants in a room | Nonplant objects, control | about 17 min | 3.5 × 6 × 2.4 m | 23 °C, 34% RH, 703 lux | Experiment (RCT) | Skin temperature, blood pressure, pain tolerance | Physiology, behavior | English | ||
| [ ] | 150 US college students (75 males and 75 females), mean age: 19.6 | 9 potted red-flowering geraniums, 9 potted non-flowering geraniums, no plants in a lab | Non-flowering plants, control | 5 min | 1.8 m | 22.4 °C | Experiment (RCT) | EEG, EDA, finger skin temperature | Physiology | American Horticultural Therapy Association | English | |
| [ ] | 146 Japanese college students (83 males and 63 females), ethnicity: Asian | 1 potted 1-m-tall plant placed in front of the participant, the same plant placed on the right-hand side of the participant, no plants in a room | Control | 15 min | 2.345 m in front of and 1.75 m at the side of the participants | 5.81 × 2.78 × 2.35 m | Experiment (RCT) | An association task, a sorting task | Cognition | English | ||
| [ ] | 66 US college students (32 males and 34 females), age: 91% from 18 to 24 | 1 potted flower arrangement (45 × 45 × 45 cm), lavender fragrance, flower and fragrance, or no plants and no fragrance in a lab | Control | 30 min | 3.5 × 2.7 × 2.4 m | 21 °C, 10.6 μmol·m ·s | Experiment (RCT) | EEG, EDA, skin temperature | Physiology | English | ||
| [ ] | 90 US college female students, mean age: 18.9 | Foliage and flowing plants, flowing plants, or no plants in a lab | Control | 5 min maximum | 1.4 m | 3.9 × 2.3 × 2.7 m | 21.7 °C, 904 lux | Experiment (RCT) | Pain tolerance, EEG, EDA, finger skin temperature | Behavior, physiology | English | |
| [ ] | 90 Japanese college students (35 males and 55 females), ethnicity: Asian | 1 potted 1.5-m-tall plant, a magazine rack put at the same location, or no plants and no magazine racks in a room | A magazine rack, control | 15 min | About 2.9 m in front of the participant | 2.78 × 5.81 × 2.35 m | Experiment (RCT) | An association task | Cognition | English | ||
| [ ] | 38 Taiwanese college students (10 males and 28 females), ethnicity: Asian | Presentation of 6 slides (office without a window view nor indoor plants, office without a window view but with indoor plants, office with a city window view but without indoor plants, office with a city window view and with indoor plants, office with a nature window view but without indoor plants, and office with a nature window view and with indoor plants) in a lab | Control | 15 s for each slide | 3 m | 7 × 5 m | 25 °C | Experiment (non-RCT) | EEG, EMG, BVP | Physiology | English | |
| [ ] | 364 Norwegian office workers, mean age: 43.1 | Presence or absence of potted plants on desks or shelves in an office | Survey (non-RCT) | Sick leave | Health | English | ||||||
| [ ] | 50 healthy Swedish people (23 males and 27 females), mean age: 39.2 | 1 potted flowering begonias ( ) approximately 22 cm high (control plant irrigated with ordinary local tap water; experiment plant irrigated with vortex-rotated local tap water) in an office | Plant irrigated with ordinary local tap water | 10 min for each plant | 5.6 × 3.0 × 2.4 m | 23–24 °C, 36–38% RH, 570–650 lux | Experiment (RCT) | Heart rate, heart rate variability, power spectral density | Physiology | The Swedish Flower Corporations | English | |
| [ ] | 90 South Korean patients who had received appendectomy (52 males and 38 females), mean age: 37.6, ethnicity: Asian | Presence or absence of 12 potted flowering plants in a ward | Control | Field experiment (RCT) | Pain killer consumption, blood pressure, body temperature, heart rate, respiratory rate | Health, physiology | English | |||||
| [ ] | 140 South Korean female high school students, ethnicity: Asian | Presence or absence of plants in 2 classrooms (accounting for 5% of the space) | Control | 14 weeks of school time | Field quasi-experiment (non-RCT, pre-post design) | Cortisol level, health | Physiology, health | English | ||||
| [ ] | 89 US sophomores | Presence or absence of plants in a classroom | Control | 1 semester of class time | Field quasi-experiment (non-RCT) | Course grade | Cognition | English | ||||
| [ ] | 76 Taiwanese junior high school students (58 males and 18 females), mean age: 13.55, ethnicity: Asian | Presence or absence of 6 potted plants (about 135 × 80 cm, having a green coverage ratio of 6%) in a classroom | Control | 12 weeks of school time | Field quasi-experiment (non-RCT) | Sick leave, misconduct | Health, behavior | English | ||||
| [ ] | 80 South Korean female patients who had received thyroidectomy, mean age: 36.2, ethnicity: Asian | Presence or absence of 12 potted flowering plants in a ward | Control | Field experiment (RCT) | Pain killer consumption, hospitalization days | Health | English | |||||
| [ ] | 34 Norwegian college students (12 males and 22 females), mean age: 24.15 | Presence or absence of 4 potted plants (2 flowering pink , 1 30-cm-tall , and 1 120-cm-tall ) in an office | Control | 60 min | 3.9 × 2.1 × 3.6 m | Experiment (RCT) | The Reading Span Task | Cognition | English | |||
| [ ] | 36 Taiwanese junior high school students (18 males and 18 females), mean age: 12.41, ethnicity: Asian | Taking care of 34 potted plants inside and outside a classroom (with a green coverage ratio of 6.3% indoors) | Control | 18 weeks of school time | Field experiment (RCT) | Examination score | Cognition | Chinese | ||||
| [ ] | 30 Chinese college students (15 males and 15 females), ethnicity: Asian | Presentation of 5 photos of vegetation landscapes and a blank in a room | Control | 2 min | 0.5 m | 7 × 4 × 3 m | 25 °C, 40% RH | Experiment (RCT) | ECG, blood pressure, heart rate, GSR, fingertip pulse | Physiology | English | |
| [ ] | 30 Chinese college students (15 males and 15 females), age: 18 to 24, ethnicity: Asian | Presentation of 12 photos of flowers and a blank in a room | Control | 2 min | 0.5 m | 7 × 4 × 3 m | 25 °C, 40% RH | Experiment (RCT) | Blood pressure, heart rate, GSR, fingertip plus | Physiology | National Key Technology Research and Development Program in China | English |
| [ ] | 29 Japanese college students (14 males and 15 females), age: 19 to 24, ethnicity: Asian | Potted L. (60 × 40 cm) of 5 different colors on a table in a room | Different colors of the plant | 1 min for each plant color | 0.5 m | Experiment (RCT) | Brain activity, eye movement | Cognition | Egyptian Ministry of Higher Education | English | ||
| [ ] | 28 Japanese undergraduate and graduate students (14 males and 14 females), mean age: 21.42, ethnicity: Asian | Placement of 1 potted plant of 3 different colors on a table in a room | Different colors of plants | 1 min for each plant color | 1.5 m | 59.4 m | 23 °C, 55% RH, 700 lux | Experiment (RCT) | Eye movement, brain activity | Cognition | Egyptian Ministry of Higher Education | English |
| [ ] | 30 South Korean college students (15 males and 15 females), mean age: 23.5, ethnicity: Asian | Placement of potted plants (60 × 40 cm) of 5 different colors on a box in a classroom | Different colors of plants | 3 min for each plant color | 1 m | 7 × 4.5 × 2.8 m | 25 °C, 70% RH, 700 lux | Experiment (RCT) | EEG | Physiology | English | |
| [ ] | Study 3: 33 British adult office workers (16 males and 17 females), mean age: 28 | Study 3: presence or absence of 8 potted plants (average height 90 cm) in an office | Control | Study 3: Field experiment (RCT) | An information management and processing task, a vigilance task | Cognition | English | |||||
| [ ] | 16 Chinese college students (8 males and 8 females), mean age: 23.5, ethnicity: Asian | Presence of potted plants of the combinations of 3 colors, 3 scents, and 3 sizes on a table in an office | Combinations of plant colors, scents, and sizes | 10–15 min | 22 °C, 41.65% RH, 0.2 ms wind velocity | Experiment (RCT) | EEG, ECG, oxyhaemoglobin saturation, fingertip blood flow, skin resistance, respiration rate | Physiology | Sciences and Technology Commission of Shanghai | English | ||
| [ ] | 24 South Korean male adults, mean age: 24.9, ethnicity: Asian | A plant transplanting task, a computer operation task on a table in a greenhouse room | A computer task | 15 min | 20.8 °C, 57.7% RH, 1365.5 lux | Experiment (RCT) | Heart rate variability, blood pressure, pulse rate | Physiology | English | |||
| [ ] | 565 Norwegian office workers | Outdoor nature contact, indoor nature contact, and outdoor view through windows | Survey (non-RCT) | Sick leave | Health | English | ||||||
| [ ] | 270 Pakistani surgical patients, ethnicity: Asian | Presence or absence of foliage plants and flower arrangements in a ward | Control | Field experiment (RCT) | Blood pressure, heart rate, respirationrate, body temperature, hospitalization days, analgesics consumption | Physiology, health | The University of Agriculture Peshawar in Pakistan | English | ||||
| [ ] | 30 Egyptian male college students, age: 22 to 37, ethnicity: African | Potted L. (60 × 40 cm) of 5 different colors on a table in a room | Different colors of the plant | 1 min for each plant color | 0.5 m | 59.4 m | 21 °C, 55% RH | Experiment (RCT) | Eye movements, brain activity | Cognition, physiology | Egyptian Ministry of Higher Education | English |
| [ ] | 5 Indonesians, ethnicity: Asian | A room with 5 potted plants and a room without plants | Control | 30 min | Experiment (non-RCT) | Heart rate, blood pressure | Physiology | Ministry of National Education in Indonesia | English | |||
| [ ] | 66 Hong Kongese college students (40 males and 26 females), mean age: 25.6, ethnicity: Asian | A basement room with plants, with a fake window, with plants and a fake window, and without plants nor a window | Control | At least 8 min | 3.3 × 2.2 × 2 m | 24 °C | Experiment (non-RCT) | EDA, a response time task | Physiology, cognition | Hong Kong Polytechnic University | English | |
| [ ] | 28 US adults (12 males and 16 females), age: 23 to 42 | Presence or absence of plants in an actual environment and a virtual one | Control | 5 min | Experiment (RCT) | Heart rate, EDA, blood pressure, a visual reaction time task, The Stroop task, a visual backward digit span task | Physiology, cognition | Campus Sustainability Innovation Fund, Harvard University Office for Sustainability | English | |||
| [ ] | 36–41 Japanese office workers, mean age: 33.95, ethnicity: Asian | Presence (3–10% green coverage ratio) or absence of plants in 2 offices | Control | 16 weeks of working hours | 132 m (321 m ), 270 m (675 m ) | Field quasi-experiment (non-RCT) | Heart rate, salivary amylase activity, critical flicker fusion frequency, fingertip pulse wave | Physiology | Grant-in-Aid for Scientific Research, Japan Society for the Promotion of Science | English | ||
| [ ] | 50 Chinese female elders with hypertension, mean age: 79.2, ethnicity: Asian | Presence or absence of 1 potted plant on a table in a room | Control | 5 min | 0.38 m | 23 °C, 40% RH, 500 lux, | Experiment (RCT) | Blood pressure, EEG | Physiology | English | ||
| [ ] | 100 Taiwanese elders, age: >65, ethnicity: Asian | Presence or absence of plants in houses | 1 year | Survey (non-RCT) | Blood pressure, heart rate | Physiology | Ministry of Science and Technology in Taiwan | English | ||||
| [ ] | 63 adult Japanese office workers (33 males and 30 females), mean age: 40.15, ethnicity: Asian | Presence or absence of 1 potted plant (15–20 cm tall, 7–10 cm wide) on the desk in an office | Control | 3 min | 1260 m | 20–24 °C, 40–50% RH, 500–700 lux | Field experiment (non-RCT, pre-post design) | Pulse rate | Physiology | English | ||
| [ ] | 30 Chinese female office workers, mean age: 29.42, ethnicity: Asian | Presence or absence of 1 potted plant with blue or purple flowers on a desk in an office | Control | 3 min | 0.4 m | 21 °C, 50% RH, 300 lux | Field quasi-experiment (non-RCT, pre-post design) | EEG, heart rate variability, skin conductance | Physiology | National Nature Science Foundation of China | English | |
| [ ] | 33 Chinese elders, age: 65 to 99, ethnicity: Asian | Combination of potted succulents (3–10 cm tall, 3 cm wide) or flower arrangement (50–60 cm tall, 5–18 cm wide) performed indoors | Flower arrangement | 25 min | Experiment (RCT) | Salivary cortisol | Physiology | National Nature Science Foundation of China | Chinese | |||
| [ ] | 34 Chinese elders with dementia (13 males and 21 females), ethnicity: Asian | With or without a treatment course of indoor horticultural activities (sowing, transplanting seedlings, succulents potting, and herbal flower potting) | Control | 30 min | Experiment (non-RCT) | Blood pressure, heart rate, ECG | Physiology | National Nature Science Foundation of China, Beijing Science and Technology Project Foundation | Chinese | |||
| [ ] | 44 Chinese elders living alone, ethnicity: Asian | Four kinds of indoor horticultural activities (sowing, transplanting seedlings, succulents potting, and herbal flower potting) | Within- participants, between-participants | 30 min | Experiment (non-RCT) | Blood pressure, heart rate, ECG | Physiology | Beijing Science and Technology Commission Green Communication Foundation | Chinese | |||
| [ ] | Study 1: 120 South Africans, mean age: 33.72, ethnicity: African | Presence of 3 potted plants, 6 plant pictures on 3 walls (80 × 80 cm), and no potted plants and plant pictures in an office | Control | 35 min | 3 × 3 m | 21 °C, 510 lux | Experiment (RCT) | A card-sorting task, a reading task | Cognition | English |
RH: relative humidity; SBP: systolic blood pressure; DBP: diastolic blood pressure; EEG: electroencephalography; EDA: electrodermal activity; EMG: electromyography; BVP: blood volume pulse; ECG: electrocardiography; GSR: galvanic skin response; RCT: randomized controlled trial; non-RCT: not randomized controlled trial.
Statistics of experimental conditions.
| Experimental Condition | Maximum | Minimum | Number of Records | |
|---|---|---|---|---|
| 1 year | 15 s. | 34 | ||
| [ ] | [ ] | |||
| Floor area | 1260 m | 7.26 m | 19 | |
| [ ] | [ ] | |||
| Volume | 675 m | 14.52 m | 14 | |
| [ ] | [ ] | |||
| 3 m | 0.38 m | 13 | ||
| [ ] | [ ] | |||
| 27 °C | 20 °C | 19 | ||
| [ ] | [ ] | |||
| 70% | 34% | 13 | ||
| [ ] | [ ] | |||
| 0.2 m·s | 1 | |||
| [ ] | ||||
| Illuminance | 1365.5 lux | 300 lux | 11 | |
| [ ] | [ ] | |||
| Quantum | 10.6 μmol·m ·s | 1 | ||
| [ ] | ||||
A total of 34 papers recorded the time during which participants were exposed to indoor plants. Among these, the longest exposure time was one year [ 76 ] and the shortest was 15 s [ 72 ]. Thirty-three papers reported the room size. The experiment room used by Toyoda et al. [ 59 ] was the largest in terms of its floor area (1260 m 2 ), and that used by Genjo et al. [ 57 ] was the largest in terms of its volume (675 m 3 ). By contrast, the room used by Kim et al. [ 77 ] was the smallest in both floor area (7.26 m 2 ) and volume (14.52 m 3 ). Among the records, only 13 reported the participant–plant distance in a room, with the greatest distance being 3 m [ 72 ] and the smallest being 0.38 m [ 60 ] ( Table 3 ).
Some records also provided data on the ambient environment in which the plants were placed. Specifically, 19 papers recorded the room temperature, with the highest being 27 °C [ 78 ] and the lowest 20 °C [ 59 ]. Humidity was reported in 13 papers, with the highest value at 70% [ 79 ] and the lowest 34% [ 80 ]. Only one record measured wind speed (0.2 m·s −1 ; [ 81 ]). Twelve records indicated lighting, of which only one adopted the quantum as the lighting unit (10.6 μmol m −2 ·s −1 ; [ 69 ]), whereas the remaining 11 used illuminance as the unit. The most intense lighting was 1365.5 lux [ 82 ], while the least intense lighting was 300 lux [ 56 ] ( Table 3 ).
Among the 42 records, only 18 indicated their funding sources. Most of the funding sources were in governmental sectors, while only two may be from stakeholders ([ 83 ], American Horticultural Therapy Association; [ 68 ], The Swedish Flower Corporations) ( Table 4 ). Funding from stakeholders might cause a conflict of interest.
3.3. Risk of Bias within Studies
Most of the included studies (90.5%) applied quasi-experimental or experimental methods. Control and experimental groups were therefore involved. In quasi-experimental research, particularly field research (11.9%), researchers were unable to assign interventions randomly to participants as is the case in clinical trials. Surveys, field quasi-experiments, and quasi-experiments, therefore, could not achieve sequence generation, which reduces the risk of bias. In addition, concealing the intervention assignment from participants was difficult because indoor plants were easily noticed in a room, resulting in lower allocation concealment ability. Similarly, blinding participants concerning their intervention was also challenging. Furthermore, the risk of incomplete data on outcomes caused by participant attrition and exclusion might exist because the included studies seldom mentioned participant attrition or exclusion.
The mean quality appraisal score of the 42 records was 17.2 points out of a possible 38, i.e., 45.3% (17.2/38 = 45.3%) of the total, indicating moderate research quality (high: 67–100%, moderate: 34–66%, low: 0–33%; [ 19 ]). The five items (of a total of 19) in the quality appraisal system for which the records included scored lowest are discussed next. First, none of the 42 papers complied with the intention-to-treat (ITT) analysis (0%) (i.e., all data were included after allocation). Additionally, the participants were not sufficiently representative because most were students (17 studies included college students, 1 study included high school students, and 2 studies included junior high school students). Only 5 papers involved general adult participants, whereas the remaining papers involved patients or office workers. The second lowest score regarding the quality appraisal system was found in the only 1 paper (2%) in which the outcome assessors were completely unaware of participant allocation. The third lowest scores were observed in the following two items of the quality appraisal system: only 2 papers (5%) reported statistical power and randomization procedure, respectively ( Table 5 and Table 6 respectively). The records exhibited desirable quality in the following items of the appraisal system: (1) all the papers (100%) included individual level analyses, (2) data collection in 39 studies (95%) was consistent, (3) a total of 37 studies (90%) provided a clear description of interventions and control, and (4) 32 papers (78%) accounted for all participants and applied statistical analysis methods appropriate for study design.
Quality appraisal of records in this study.
| Quality Indicators | [ ] | [ ] | [ ] | [ ] | [ ] | [ ] | [ ] | |
|---|---|---|---|---|---|---|---|---|
| Power calculation reported | No | No | No | No | No | No | No | |
| Inclusion/exclusion criteria reported | No | No | No | No | No | No | No | |
| Individual level allocation | No | Yes | NA | Yes | Yes | Yes | Yes | |
| Random allocation to groups/condition/order | Yes | NA | Yes | Yes | Yes | Yes | ||
| Randomization procedure appropriate | Unclear | NA | Unclear | Unclear | Unclear | Unclear | ||
| Groups similar (sociodemographic) | Unclear | Unclear | Unclear | Yes | Yes | Unclear | Yes | |
| Group balanced at baseline | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Yes | |
| Participants blind to research question | Yes | Unclear | Unclear | Unclear | Unclear | |||
| Clear description of intervention and control | Yes | Yes | NA | Yes | Yes | Yes | Yes | |
| Consistency of intervention (within and between groups) | Yes | No | NA | Yes | Yes | Yes | No | |
| Outcome assessors blind to group allocation | Unclear | Unclear | Unclear | Unclear | ||||
| Baseline measures taken before the intervention | Yes | Unclear | NA | Yes | Yes | No | Yes | |
| Consistency of data collection | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| All outcomes reported (means and SD/SE) | No | Yes | No | No | No | Yes | No | |
| All participants accounted for (i.e., losses/exclusions) | Yes | Yes | Yes | Yes | Yes | No | Yes | |
| ITT analysis conducted (all data included after allocation) | Unclear | Unclear | NA | Unclear | Unclear | No | Unclear | |
| Individual level analysis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Statistical analysis methods appropriate for study design | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Sample representative of target population | No | No | No | No | No | No | No | |
| Total number of points (out of possible 38) | 20 | 18 | 8 | 20 | 20 | 16 | 22 | |
| Quality rating as percent | 52.6 (M) | 47.4 (M) | 21.1 (L) | 52.6 (M) | 52.6 (M) | 42.1 (M) | 57.9 (M) | |
| Responded to query about “uncertain” ratings | Yes | Yes | NA | No | NA | Yes | ||
| ] | ] | ] | ] | ] | ] | ] | ||
| Power calculation reported | No | No | No | No | No | No | No | |
| Inclusion/exclusion criteria reported | No | No | Yes | No | No | Yes | No | |
| Individual level allocation | Yes | Yes | Yes | NA | Yes | Yes | No | |
| Random allocation to groups/condition/order | Yes | Yes | Unclear | NA | Yes | Yes | No | |
| Randomization procedure appropriate | Unclear | Unclear | Unclear | NA | Unclear | Unclear | NA | |
| Groups similar (sociodemographic) | Yes | Unclear | Yes | Unclear | Unclear | Unclear | Yes | |
| Group balanced at baseline | Unclear | Unclear | Yes | Unclear | Unclear | Unclear | Yes | |
| Participants blind to research question | Unclear | Unclear | Yes | Unclear | Yes | Unclear | ||
| Clear description of intervention and control | Yes | Yes | Yes | NA | Yes | Yes | Partial | |
| Consistency of intervention (within and between groups) | No | No | No | NA | Yes | Yes | No | |
| Outcome assessors blind to group allocation | Unclear | Unclear | NA | Unclear | Unclear | Unclear | ||
| Baseline measures taken before the intervention | Yes | Yes | No | NA | Yes | No | Yes | |
| Consistency of data collection | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| All outcomes reported (means and SD/SE) | No | Yes | Yes | No | Yes | No | No | |
| All participants accounted for (i.e., losses/exclusions) | Yes | Yes | Yes | No | Yes | Yes | No | |
| ITT analysis conducted (all data included after allocation) | Unclear | Unclear | Unclear | NA | Unclear | Unclear | No | |
| Individual level analysis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Statistical analysis methods appropriate for study design | Yes | Yes | No | Yes | Yes | Yes | No | |
| Sample representative of target population | No | No | No | No | No | No | No | |
| Total number of points (out of possible 38) | 18 | 20 | 18 | 8 | 20 | 20 | 11 | |
| Quality rating as percent | 47.4 (M) | 52.6 (M) | 47.4 (M) | 21.1 (L) | 52.6 (M) | 52.6 (M) | 28.9 (L) | |
| Responded to query about “uncertain” ratings | NA | Yes | No | No | ||||
| ] | ] | ] | ] | ] | ] | ] | ||
| Power calculation reported | No | No | No | No | No | No | No | |
| Inclusion/exclusion criteria reported | No | Yes | Yes | No | No | Yes | Yes | |
| Individual level allocation | No | No | Yes | Yes | Yes | Yes | Yes | |
| Random allocation to groups/condition/order | No | No | Yes | Yes | Yes | Yes | Yes | |
| Randomization procedure appropriate | NA | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | |
| Groups similar (sociodemographic) | Partial | Partial | Unclear | Unclear | Yes | Yes | Yes | |
| Group balanced at baseline | Unclear | Unclear | Unclear | Partial | Unclear | Yes | Yes | |
| Participants blind to research question | Unclear | Yes | Yes | Yes | Yes | Unclear | Unclear | |
| Clear description of intervention and control | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Consistency of intervention (within and between groups) | Yes | Yes | Yes | Yes | No | No | No | |
| Outcome assessors blind to group allocation | Unclear | No | Unclear | Unclear | No | Unclear | Unclear | |
| Baseline measures taken before the intervention | No | Yes | No | Yes | Yes | Yes | Yes | |
| Consistency of data collection | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| All outcomes reported (means and SD/SE) | Yes | No | No | No | No | Yes | Yes | |
| All participants accounted for (i.e., losses/exclusions) | No | Yes | Yes | Yes | Yes | Yes | Yes | |
| ITT analysis conducted (all data included after allocation) | No | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | |
| Individual level analysis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Statistical analysis methods appropriate for study design | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Sample representative of target population | No | No | No | No | No | No | No | |
| Total number of points (out of possible 38) | 13 | 19 | 20 | 21 | 20 | 24 | 24 | |
| Quality rating as percent | 34.2 (M) | 50.0 (M) | 52.6 (M) | 55.3 (M) | 52.6 (M) | 63.2 (M) | 63.2 (M) | |
| Responded to query about “uncertain” ratings | No | No | No | |||||
| ] | ] | ] | ] | ] | ] | ] | ||
| Power calculation reported | No | No | No | Study 3: No | Yes | No | No | |
| Inclusion/exclusion criteria reported | Yes | Yes | Yes | Study 3: No | Yes | Yes | Yes | |
| Individual level allocation | Yes | Yes | Yes | Study 3: No | No | Yes | NA | |
| Random allocation to groups/condition/order | Yes | Yes | Yes | Study 3: Yes | Unclear | Yes | NA | |
| Randomization procedure appropriate | Unclear | Unclear | Unclear | Study 3: Unclear | Unclear | Unclear | NA | |
| Groups similar (sociodemographic) | Yes | Yes | Yes | Study 3: Unclear | Yes | Yes | Unclear | |
| Group balanced at baseline | Yes | Yes | Yes | Study 3: Unclear | Yes | Yes | Unclear | |
| Participants blind to research question | Unclear | Unclear | Study 3: | Unclear | No | Unclear | ||
| Clear description of intervention and control | Yes | Yes | Yes | Study 3: Yes | Yes | Yes | NA | |
| Consistency of intervention (within and between groups) | No | No | No | Study 3: No | No | Yes | NA | |
| Outcome assessors blind to group allocation | Unclear | Unclear | Study 3: | Unclear | Unclear | Unclear | ||
| Baseline measures taken before the intervention | No | No | No | Study 3: No | No | Yes | NA | |
| Consistency of data collection | Yes | Yes | Yes | Study 3: Yes | Yes | Yes | Yes | |
| All outcomes reported (means and SD/SE) | No | No | Yes | Study 3: No | No | No | No | |
| All participants accounted for (i.e., losses/exclusions) | Yes | Yes | Yes | Study 3: Yes | Yes | Yes | No | |
| ITT analysis conducted (all data included after allocation) | Unclear | Unclear | Unclear | Study 3: Unclear | Unclear | Unclear | NA | |
| Individual level analysis | Yes | Yes | Yes | Study 3: Yes | Yes | Yes | Yes | |
| Statistical analysis methods appropriate for study design | Yes | Yes | No | Study 3: Yes | No | Yes | Yes | |
| Sample representative of target population | No | No | No | Study 3: No | No | No | No | |
| Total number of points (out of possible 38) | 20 | 20 | 20 | Study 3: 14 | 16 | 24 | 8 | |
| Quality rating as percent | 52.6 (M) | 52.6 (M) | 52.6 (M) | Study 3: 36.8 (M) | 42.1 (M) | 63.2 (M) | 21.1 (L) | |
| Responded to query about “uncertain” ratings | Yes | No | No | Yes | ||||
| ] | ] | ] | ] | ] | ] | ] | ||
| Power calculation reported | No | No | No | No | No | No | No | |
| Inclusion/exclusion criteria reported | Yes | Yes | No | No | Yes | No | Yes | |
| Individual level allocation | Yes | Yes | Unclear | Yes | Yes | No | Yes | |
| Random allocation to groups/condition/order | Yes | Yes | Unclear | Unclear | Yes | No | Yes | |
| Randomization procedure appropriate | Unclear | Yes | Unclear | Unclear | Unclear | NA | Unclear | |
| Groups similar (sociodemographic) | Unclear | Unclear | Yes | Unclear | Yes | Unclear | Unclear | |
| Group balanced at baseline | Unclear | Unclear | Yes | Unclear | Yes | Unclear | Unclear | |
| Participants blind to research question | Unclear | No | Unclear | Unclear | Unclear | Unclear | Unclear | |
| Clear description of intervention and control | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Consistency of intervention (within and between groups) | Yes | No | No | No | Yes | No | Yes | |
| Outcome assessors blind to group allocation | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | |
| Baseline measures taken before the intervention | No | No | No | Yes | Yes | Yes | Partial | |
| Consistency of data collection | Yes | Yes | Yes | Yes | Yes | No | Yes | |
| All outcomes reported (means and SD/SE) | No | No | No | No | No | No | No | |
| All participants accounted for (i.e., losses/exclusions) | Yes | Yes | Yes | Yes | No | No | Yes | |
| ITT analysis conducted (all data included after allocation) | Unclear | Unclear | Unclear | Unclear | No | Unclear | Unclear | |
| Individual level analysis | Yes | Yes | Unclear | Yes | Yes | Yes | Yes | |
| Statistical analysis methods appropriate for study design | No | No | Unclear | Yes | Yes | No | Yes | |
| Sample representative of target population | No | No | No | No | No | No | No | |
| Total number of points (out of possible 38) | 16 | 16 | 10 | 14 | 22 | 6 | 19 | |
| Quality rating as percent | 42.1 (M) | 42.1 (M) | 26.3 (L) | 36.8 (M) | 58.9 (M) | 15.8 (L) | 50.0 (M) | |
| Responded to query about “uncertain” ratings | ||||||||
| ] | ] | ] | ] | ] | ] | ] | ||
| Power calculation reported | No | No | No | No | No | No | Yes | |
| Inclusion/exclusion criteria reported | Yes | No | Yes | Yes | No | Yes | No | |
| Individual level allocation | NA | No | Yes | No | Unclear | Unclear | Yes | |
| Random allocation to groups/condition/order | NA | No | No | Yes | Unclear | Unclear | Yes | |
| Randomization procedure appropriate | NA | NA | NA | Unclear | Unclear | Unclear | Unclear | |
| Groups similar (sociodemographic) | Unclear | Yes | Yes | Unclear | Yes | Unclear | Unclear | |
| Group balanced at baseline | Unclear | Yes | Yes | Unclear | Yes | Unclear | Unclear | |
| Participants blind to research question | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Yes | |
| Clear description of intervention and control | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Consistency of intervention (within and between groups) | Yes | No | No | Yes | No | No | No | |
| Outcome assessors blind to group allocation | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | |
| Baseline measures taken before the intervention | No | Yes | Yes | Yes | Yes | Yes | No | |
| Consistency of data collection | Yes | No | Yes | Yes | Yes | Yes | Yes | |
| All outcomes reported (means and SD/SE) | Yes | No | No | Yes | Yes | Yes | Yes | |
| All participants accounted for (i.e., losses/exclusions) | Yes | Yes | Yes | No | Yes | No | Yes | |
| ITT analysis conducted (all data included after allocation) | NA | Unclear | Unclear | No | Unclear | No | Unclear | |
| Individual level analysis | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| Statistical analysis methods appropriate for study design | Yes | No | Yes | Yes | Yes | Yes | Yes | |
| Sample representative of target population | No | No | No | No | No | No | No | |
| Total number of points (out of possible 38) | 16 | 12 | 20 | 18 | 18 | 14 | 20 | |
| Quality rating as percent | 42.1 (M) | 31.6 (L) | 52.6 (M) | 47.4 (M) | 47.4 (M) | 36.8 (M) | 52.6 (M) | |
| Responded to query about “uncertain” ratings | ||||||||
ITT: intention to treatment; Yes = 2; Partial (Pa.) = 1; No = 0; Unclear (Un) = 0; NA = criterion inapplicable to this study design; any changes made after consultation with study authors are highlighted in boldface. Appraisal quality: High (H): 67–100%, Moderate (M): 34–66%, Low (L): 0–33% [ 19 ].
Statistics of quality appraisal of records in this study.
| Yes | Partial | No | Unclear | NA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Frequency | (%) | Frequency | (%) | Frequency | (%) | Frequency | (%) | Frequency | (%) | |
| Power Calculation Reported | 2 | 5 | 0 | 0 | 39 | 95 | 0 | 0 | 0 | 0 |
| Inclusion/exclusion Criteria Reported | 20 | 49 | 0 | 0 | 21 | 51 | 0 | 0 | 0 | 0 |
| Individual Level Allocation | 26 | 63 | 0 | 0 | 8 | 20 | 3 | 7 | 4 | 10 |
| Random Allocation to Groups/Condition/Order | 25 | 61 | 0 | 0 | 6 | 15 | 6 | 15 | 4 | 10 |
| Randomization Procedure Appropriate | 2 | 5 | 0 | 0 | 0 | 0 | 30 | 73 | 9 | 22 |
| Groups Similar (Sociodemographic) | 19 | 46 | 2 | 5 | 0 | 0 | 20 | 49 | 0 | 0 |
| Group Balanced at Baseline | 15 | 37 | 1 | 2 | 0 | 0 | 25 | 61 | 0 | 0 |
| Participants Blind to Research Question | 11 | 27 | 0 | 0 | 3 | 7 | 27 | 66 | 0 | 0 |
| Clear Description of Intervention and Control | 37 | 90 | 1 | 2 | 0 | 0 | 0 | 0 | 3 | 7 |
| Consistency of Intervention (within and between groups) | 16 | 39 | 0 | 0 | 22 | 54 | 0 | 0 | 3 | 7 |
| Outcome Assessors Blind to Group Allocation | 1 | 2 | 0 | 0 | 6 | 15 | 33 | 80 | 1 | 2 |
| Baseline Measures Taken before the Intervention | 22 | 54 | 1 | 2 | 14 | 34 | 1 | 2 | 3 | 7 |
| Consistency of Data Collection | 39 | 95 | 0 | 0 | 2 | 5 | 0 | 0 | 0 | 0 |
| All Outcomes Reported (Means and SD/SE) | 14 | 34 | 0 | 0 | 27 | 66 | 0 | 0 | 0 | 0 |
| All Participants Accounted for (i.e., losses/exclusions) | 32 | 78 | 0 | 0 | 9 | 22 | 0 | 0 | 0 | 0 |
| ITT Analysis Conducted (all data included after allocation) | 0 | 0 | 0 | 0 | 6 | 15 | 31 | 76 | 4 | 10 |
| Individual Level Analysis | 40 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Statistical Analysis Methods Appropriate for Study Design | 32 | 78 | 0 | 0 | 8 | 20 | 1 | 2 | 0 | 0 |
| Sample Representative of Target Population | 0 | 0 | 0 | 0 | 41 | 100 | 0 | 0 | 0 | 0 |
3.4. Results of Individual Studies
The research outcomes of each study for the systematic review are summarized in Table 7 . In brief, the systematic review concluded that indoor plants, in general, affect participants’ functions positively, particularly their physiology and cognition. Regarding physiological functions, participants exhibited greater benefits in a room with plants than in a room without plants in relation to lower blood pressure [ 60 , 61 , 63 , 76 , 78 , 82 ], lower electrodermal activity (EDA) [ 69 , 83 , 85 ], lower electroencephalography (EEG) α and β waves [ 56 , 69 , 72 , 81 , 83 ], lower heart rate [ 59 , 61 , 62 , 63 , 68 , 76 , 91 , 93 ], and lower respiration rate and body temperature [ 61 ].
Summary of the outcomes of the records.
| Source | Outcomes |
|---|---|
| [ ] | When conducting a computer task, participants had a smaller SBP increase with the presence of plants than without plants. After accomplishing the task, the participants also exhibited a faster SBP decrease when plants were present than when plants were absent. Participants’ reaction time was 12% faster when plants were present than when they were absent. |
| [ ] | Participants had the lowest productivity when the office was furnished with 22 potted plants, whereas the highest productivity was observed when no plants were present. |
| [ ] | Participants had a significantly lower search error rate with indoor greening than without indoor greening. |
| [ ] | The percentage of participants putting their hands in ice water for more than 5 min was higher with the presence of plants than without plants. |
| [ ] | Female participants’ decreases in EEG β waves and EDA were significantly faster when red-flowering geraniums were present than when flowerless geraniums were present and when plants were absent. |
| [ ] | Male participants had a lower score in the association task than their female counterparts when plants were absent, whereas female participants had higher scores on the sorting task regardless of the presence or absence of plants. |
| [ ] | Female participants’ EEG β waves and EDA were significantly lower when flower arrangements were present than when flower arrangements were absent. |
| [ ] | Participants’ time of hand immersion in ice water was significantly longer when green-leaf and flowering plants were simultaneously present than when only green-leaf plants or flowering plants were in the room and when plants were not in the room. Participants’ EDA was significantly lower when the plants were in the room than when the plants were not in the room. |
| [ ] | Female participants showed significantly higher scores of the association task than male participants in the three interventions. Female participants had significantly higher scores of the association task when plants were present than when the magazine-rack was present. |
| [ ] | Participants had the greatest effect of EEG β waves when viewing the slide of the office with a nature window view and indoor plants than other slides. |
| [ ] | A weak but significant correlation was observed between the number of potted plants and sick leave days in the workplace. |
| [ ] | The increased humidity of the indoor potted plants improved the vagus-induced sympathovagal balance of the heart of the participant. |
| [ ] | Participants’ frequency of pain killer consumption, SBP, and heart rate were significantly lower when plants were in the room than when plants were not in the room. |
| [ ] | Participants’ frequency of visiting the school infirmary was significantly lower when plants were in the room than when plants were not in the room. |
| [ ] | Participants’ grade point averages wer significantly higher when plants were present than when plants were absent. |
| [ ] | Participants’ sick leave hours and misconduct were significantly less when plants were present than when plants were absent. |
| [ ] | Participants’ frequency of pain killer use and hospitalization days were significantly lower when plants were in the room than when plants were not in the room. |
| [ ] | Participants’ attention improved significantly from the baseline to after the proofreading task was completed when plants were present, whereas no improvement was noted when plants were absent. |
| [ ] | Participants who took care of plants had greater academic achievement than those who did not. |
| [ ] | Red, yellow, and green plants significantly reduced participants’ DBP and fingertip pulse. Red, purple, and yellow plants significantly reduced participants’ fingertip pulse. Changes in fingertip pulse were more significant in male participants than in female participants. |
| [ ] | Except for yellow African daisies, the other flowers significantly reduced participants’ SBP. Pink and white African daisies, pink and white carnations, and pink and white roses significantly reduced participants’ DBP. |
| [ ] | Male participants spent significantly more time looking at white L. than at the dark green variety. Female participants had a greater frequency of looking at yellow-green plants than looking at dark green and green-white plants. |
| [ ] | Male participants spent significantly more time looking at green plants than at red-green ones. The number of fixings at red–green plants was greater than at green and white–green plants. Female participants spent significantly more time looking at green and red–green plants and with greater frequency than green–white plants. |
| [ ] | Relative to green plants with white, yellow, pink, and red flowers, green-leaf plants resulted in a greater increase in participants’ relative slow α power, relative fast α power, relative low β power, and relative moderate β power spectra. By contrast, green-leaf plants with yellow flowers increased participants’ relative θ power spectrum. |
| [ ] | Participants spent less time completing the vigilance and information processing tasks when plants were present than when plants were absent. |
| [ ] | Participants had a significantly higher δ waves and significantly lower α and β waves when plants were present than when plants were absent. |
| [ ] | After transplanting plants, participants had a significantly lower DBP than their counterparts did after a computer operation task. |
| [ ] | The indoor nature contact during work was significantly negatively correlated with sick leave days. |
| [ ] | The percentage of patients with stable blood pressure, heart rate, respiration rate, and body temperature was significantly higher in the ward with plants than in the one without plants. These patients also received a significantly lower dose of pain killers and had significantly shorter hospitalization. |
| [ ] | Yellow–green L. received more attention than did the plants of other colors. |
| [ ] | Participants had lower heart rate in the room when the plants were present than when the plants were not present. |
| [ ] | Participants had a significantly faster reaction rate when plants were present than when plants were absent. |
| [ ] | In both the actual and virtual environments with plants, participants exhibited greater changes in SBP, DBP, and EDA than in the plantless environment. They also had greater performance in the visual backward digit span task in the plant setting. |
| [ ] | Participants had the least flicker fusion frequency (eye fatigue) when flowering plants were provided than with other plants and controls. |
| [ ] | Participants had significantly lower SBP and a significant increase in the amplitude of high β waves when plants were present than when plants were absent. |
| [ ] | Participants without houseplants had significantly higher SBP and heart rate than those with houseplants. |
| [ ] | Participants had a significantly greater proportion of significantly decreased pulse rate when the plant was present than when the plant was absent. |
| [ ] | Participants had a significant increase in α relative waves in the prefrontal and occipital lobes and in parasympathetic nervous activity when the plant was present than when the plant was absent. |
| [ ] | There were significant differences between the two horticultural activities and between the pretest and the posttest. |
| [ ] | There were significant differences between the experimental and the control groups in heart rate variability (standard deviation of the NN intervals, root mean square of the successive differences, low frequency, high frequency, and low frequency/high frequency). Within the treatment, male participants’ standard deviation of the NN intervals was significantly different between sowing and transplanting seedlings. |
| [ ] | Participants had a significantly lower heart rate after sowing, transplanting seedlings, and potting succulents. Among the four kinds of horticultural activities, sowing yielded the greatest heart rate reduction while herbal flower potting was the worst. |
| [ ] | Participants had significantly fewer errors and faster time of task completion when the plants and pictures were present than when they were absent. |
SBP: systolic blood pressure; DBP: diastolic blood pressure; EEG: electroencephalography; EDA: electrodermal activity.
Regarding cognitive functions, when indoor plants were present, participants exhibited higher academic achievement [ 66 , 86 ] and better performance in various cognitive tasks [ 58 , 71 , 75 , 77 , 78 , 84 , 87 , 94 ]. In health-related functions, with exposure to indoor plants, participants less frequently took sick leave [ 54 , 55 , 65 , 67 ], consumed fewer pain killers [ 61 , 63 , 64 ], and had fewer hospitalization days [ 64 ] than participants in environments where indoor plants were absent. In behavioral functions, participants presented greater pain tolerance of putting hands in cold water [ 80 , 85 ] and less misconduct [ 65 ] when indoor plants were in the room than when indoor plants were not in the room.
3.5. Synthesis of Results
The data for the meta-analyses included only the participants’ physiological functions (i.e., diastolic blood pressure (DBP), EEG α and β waves) and cognitive functions (i.e., attention, academic achievement, and response time) because at least two studies are needed to conduct the meta-analyses. Given that the number of the records of each of the function categories was small, randomized control trials and non-randomized studies of interventions were included for the meta-analyses. Moreover, various interventions of indoor plants regardless of species, type, quantity, exposure time, and distance to participants were dichotomized as groups with plants and groups without plants.
Three papers examining the influence of indoor plants on DBP, which was measured by sphygmomanometers measured in mmHg, were included for the meta-analysis ( Table 8 ). In total, 248 participants were evenly exposed to conditions either with plants or without plants. Lee et al. [ 82 ] recruited only male adults in South Korea, whereas Hassan et al. [ 60 ] recruited only female older adults with high blood pressure in China. Chen et al. [ 76 ] surveyed male and female elders in Taiwan six times over one year. Both Lee et al. [ 82 ] and Hassan et al. [ 60 ] randomly assigned their participants to different groups, while Chen et al. [ 76 ] did not. All three papers were appraised as having moderate research quality.
Original data of the studies examining the influence of indoor plants on DBP.
| Study | Study Design | Appraisal Quality | Without Plant | With Plant | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| [ ] | Experiment (RCT) | Moderate | 24 | 71.75 | 0.78 | 24 | 65.26 | 0.69 |
| [ ] | Experiment (RCT) | Moderate | 50 | 68.2 | 5.77 | 50 | 67.3 | 9.05 |
| [ ] | Survey (non-RCT) | Moderate | 300 | 74.20 | 6.20 | 300 | 70.10 | 6.00 |
The heterogeneity test of the three studies focusing on DBP revealed a significant difference ( p < 0.05) with I 2 = 97.554%, confirming high heterogeneity among the studies. A random-effect model was therefore applied. Given that the standard deviation (SD) of one study was much smaller than that of the other two, SMD, rather than MD, was adopted here. The pooled effect size (SMD) was −2.526 with a 95% confidence interval ranging between −4.142 and −0.909. The results indicated that the group with plants had significantly ( p = 0.002) lower DBP values than the group without plants ( Table 9 ). The relative weight of both the Hassan et al. [ 60 ] and Chen et al. [ 76 ] studies was about 37.00%, and that of Lee et al. [ 82 ] was 25.29% ( Figure 2 ).

Forest plot of studies on the influence of indoor plants on DBP [ 60 , 76 , 82 ].
Heterogeneity test results of studies on the influence of indoor plants on DBP.
| Model | Number of Studies | Pooled Effect Size | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
| Effect Size | Standard Error | -Value | Q-Value | df (Q) | -Value | I-Squared | ||
| Fixed | 3 | −0.644 | 0.077 | <0.001 | 81.782 | 2 | <0.001 | 97.554 |
| Random | 3 | −2.526 | 0.825 | 0.002 | ||||
3.7. EEG α Waves
Three papers examining the influence of indoor plants on EEG α waves, which was measured by brain activity instruments with Hertz as the unit of measurement, were included for the meta-analysis ( Table 10 ). The studies had a total of 200 participants. Among them, 85 were in the control group (without plants) and 115 in the experimental group (with plants). Chang and Chen [ 72 ] recruited college students in Taiwan and Qin et al. [ 81 ] recruited college students in China, whereas Elasdek and Liu [ 56 ] recruited only female office workers in China. Chang and Chen [ 72 ] and Elasdek and Liu [ 56 ] did not randomly assign their participants to different groups, while Qin et al. [ 81 ] did. These three papers were appraised as having moderate research quality.
Original data of the studies examining the influence of indoor plants on EEG α waves.
| Study | Study Design | Appraisal Quality | Without Plant | With Plant | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| [ ] | Experiment (non-RCT) | Moderate | 38 | 0.130 | 0.210 | 38 | 0.090 | 0.170 |
| [ ] | Experiment (RCT) | Moderate | 17 | 0.043 | 0.020 | 17 | 0.112 | 0.027 |
| [ ] | Field quasi- experiment (non-RCT) | Moderate | 30 | 0.160 | 0.054 | 60 | 0.210 | 0.054 |
The heterogeneity test of the three studies investigating the influence of indoor plants on EEG α waves revealed a significant difference ( p < 0.05), with I 2 = 94.488%, confirming high heterogeneity among the studies. A random-effect model was therefore adopted. The pooled effect size (MD) was 1.140, and the 95% confidence interval ranged from −0.260 to 2.540. The results indicated that the group with plants had greater EEG α waves than the group without plants, but the difference was nonsignificant ( p = 0.110) ( Table 11 ). The relative weight of both the Chang and Chen [ 72 ] and Elasdek and Liu [ 56 ] studies was about 34.6%, and that of Qin et al. [ 81 ] was 30.72% ( Figure 3 ).

Forest plot of studies on the influence of indoor plants on EEG α waves [ 56 , 72 , 81 ].
Heterogeneity test results of studies on the influence of indoor plants on EEG α waves.
| Model | Number of Studies | Pooled Effect Size | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
| Effect Size | Standard Error | -Value | Q-Value | df (Q) | -Value | I-Squared | ||
| Fixed | 3 | 0.605 | 0.156 | <0.001 | 36.285 | 2 | <0.001 | 94.488 |
| Random | 3 | 1.140 | 0.714 | 0.110 | ||||
3.8. EEG β Waves
Only two papers examining the influence of indoor plants on EEG β waves, which was measured in Hertz by brain activity instruments, were included in this meta-analysis ( Table 12 ). In total, 110 participants were evenly assigned to groups either with plants or without plants. Chang and Chen [ 72 ] recruited college students in Taiwan and Qin et al. [ 81 ] recruited college students in China. Chang and Chen [ 72 ] did not randomly assign their participants to different groups, while Qin et al. [ 81 ] did. Both papers were appraised as having moderate research quality.
Original data of the studies examining the influence of indoor plants on EEG β waves.
| Study | Study Design | Appraisal Quality | Without Plant | With Plant | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| [ ] | Experiment (non-RCT) | Moderate | 38 | 0.160 | 0.240 | 38 | 0.120 | 0.220 |
| [ ] | Experiment (RCT) | Moderate | 17 | 0.051 | 0.046 | 17 | 0.214 | 0.057 |
The heterogeneity test of the two studies investigating the influence of indoor plants on EEG β waves revealed a significant difference ( p < 0.05), with I 2 = 97.133%, confirming high heterogeneity between the studies. A random-effect model was therefore adopted. The pooled effect size (MD) was 1.455, and the 95% confidence interval ranged from −1.799 to 4.709. Though the results indicated that the group with plants had greater EEG β waves than the group without plants, the difference was not significant ( p = 0.381) ( Table 13 ). The relative weight of both the Chang and Chen [ 72 ] and Qin et al. [ 81 ] studies was about equal, at 50.95% and 49.05%, respectively ( Figure 4 ).

Forest plot of studies on the influence of indoor plants on EEG β waves [ 72 , 81 ].
Heterogeneity test results of studies on the influence of indoor plants on EEG β waves.
| Model | Number of Studies | Pooled Effect Size | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
| Effect Size | Standard Error | -Value | Q-Value | df (Q) | -Value | I-Squared | ||
| Fixed | 2 | 0.381 | 0.210 | 0.069 | 34.885 | 1 | <0.001 | 97.133 |
| Random | 2 | 1.455 | 1.660 | 0.381 | ||||
3.9. Attention
Three papers examining the influence of indoor plants on attention, which was measured by various cognitive tasks with the unit of measurement as performance scores, were included for the meta-analysis ( Table 14 ). In total, 177 participants were randomly assigned to different groups. Because Larsen et al. [ 53 ] divided the participants into two experimental groups (with a high or moderate number of plants) and one control group (without plants), there were 76 participants and 101 participants in the control and experimental groups, respectively. Larsen et al. [ 53 ] recruited participants in the United States, Yin et al. [ 75 ] recruited adults in the United States, and Shibata and Suzuki [ 71 ] recruited college students in Japan. All three papers were appraised as having moderate research quality.
Original data of the studies examining the influence of indoor plants on attention.
| Study | StudyDesign | Appraisal Quality | Without Plant | With Plant | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| [ ]_1 | Experiment (RCT) | Moderate | 28 | 43.55 | 6.76 | 27 | 40.28 | 6.94 |
| [ ]_2 | Experiment (RCT) | Moderate | 28 | 43.55 | 6.76 | 26 | 38.24 | 8.64 |
| [ ] | Experiment (RCT) | Moderate | 18 | 64.67 | 20.08 | 18 | 78.77 | 21.89 |
| [ ] | Experiment (RCT) | Moderate | 30 | 4.69 | 1.18 | 30 | 5.29 | 1.13 |
The heterogeneity test of the three studies (one with two experimental groups) investigating the influence of indoor plants on attention revealed a significant difference ( p < 0.05), with I 2 = 82.088%, confirming high heterogeneity among the studies. A random-effect model was therefore adopted. The pooled effect size (SMD) was −0.005, and the 95% confidence interval ranged from −0.671 to 0.661. The results indicated that the group with plants had lower attention than the group without plants. The difference, however, was not significant ( p = 0.988) ( Table 15 ). The relative weight of the three studies was relatively similar, ranging from 25.85% to 23.32% ( Figure 5 ).

Forest plot of studies on the influence of indoor plants on attention [ 53 , 71 , 75 ].
Heterogeneity test results of studies on the influence of indoor plants on attention.
| Model | Number of Studies | Pooled Effect Size | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
| Effect Size | Standard Error | -Value | Q-Value | df (Q) | -Value | I-Squared | ||
| Fixed | 4 | −0.038 | 0.143 | 0.789 | 16.749 | 3 | 0.001 | 82.088 |
| Random | 4 | −0.005 | 0.340 | 0.988 | ||||
3.10. Academic Achievement
Only two papers examining the influence of indoor plants on academic achievement, which was measured by course grades and examination scores, were included for the meta-analysis ( Table 16 ). The studies had a total of 119 participants. Among these, 58 were in the control group (without plants) and 61 in the experimental group (with plants). Doxey et al. [ 86 ] recruited sophomores in the United States, who were not randomly assigned to groups. Han and Hung [ 66 ] recruited students from a junior high school in Taiwan, who were randomly assigned to groups. The study of Doxey et al. [ 86 ] was appraised as having low research quality, while that of Han and Hung [ 66 ] was appraised as having moderate research quality.
Original data of the studies examining the influence of indoor plants on academic achievement.
| Study | Study Design | Appraisal Quality | Without Plant | With Plant | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| [ ] | Field quasi-experiment (non-RCT) | Low | 39 | 2.62 | 0.847 | 44 | 3.14 | 0.795 |
| [ ] | Field experiment (RCT) | Moderate | 19 | 0.133 | 0.009 | 17 | 0.154 | 0.098 |
The heterogeneity test of the two studies investigating the influence of indoor plants on academic achievement revealed no significant difference ( p > 0.05), with I 2 = 0%, confirming low heterogeneity between the studies. A fixed-effect model was therefore applied. The pooled effect size (SMD) was 0.534, and the 95% confidence interval ranged from 0.167 to 0.901. The results indicated that the group with plants had significantly higher academic achievement ( p = 0.004) than the group without plants ( Table 17 ). The relative weight of Doxey et al. [ 86 ] was 68.95% and that of Han and Hung [ 66 ] was 31.05% ( Figure 6 ).

Forest plot of studies on the influence of indoor plants on academic achievement [ 66 , 86 ].
Heterogeneity test results of studies on the influence of indoor plants on academic achievement.
| Model | Number of Studies | Pooled Effect Size | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
| Effect Size | Standard Error | -Value | Q-Value | df (Q) | -Value | I-Squared | ||
| Fixed | 2 | 0.534 | 0.187 | 0.004 | 0.639 | 1 | 0.424 | 0.000 |
| Random | 2 | 0.534 | 0.187 | 0.004 | ||||
3.11. Response Time
Three papers examining the influence of indoor plants on response time, which was measured by various tasks with the unit of measurement as seconds or milliseconds, were included for the meta-analysis ( Table 18 ). These studies had a total of 749 participants. Among them, 374 participants were in the control group (without plants) and 375 in the experimental group (with plants). Nieuwenhuis et al. [ 58 ] recruited adult office workers in the United Kingdom, Kim et al. [ 77 ] recruited college students in Hong Kong, and Thatcher et al. [ 94 ] recruited adults in South Africa. Nieuwenhuis et al. [ 58 ] and Thatcher et al. [ 94 ] randomly assigned their participants to different groups, while Kim et al. [ 77 ] did not. All three papers were appraised as having moderate research quality.
Original data of the studies examining the influence of indoor plants on response time.
| Study | Study Design | Appraisal Quality | Without Plant | With Plant | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||||
| [ ] | Field experiment (RCT) | Moderate | 17 | 20.390 | 5.870 | 16 | 17.390 | 3.850 |
| [ ] | Experiment (non-RCT) | Moderate | 317 | 289.900 | 51.115 | 319 | 286.100 | 40.377 |
| [ ] | Experiment (RCT) | Moderate | 40 | 1228.000 | 258.720 | 40 | 738.650 | 186.180 |
The heterogeneity test of the three studies investigating the influence of indoor plants on response time revealed a significant difference ( p < 0.05), with I 2 = 96.144%, confirming high heterogeneity among the studies. A random-effect model was therefore adopted. Given that great differences existed between the original data, SMD, rather than MD, was adopted. The pooled effect size (SMD) was −0.939, and the 95% confidence interval ranged from −2.208 to 0.401. The results indicated that the group with plants had less response time than did the group without plants. However, the difference was not significant ( p = 0.170) ( Table 19 ). The relative weight of the three studies was relatively similar, ranging from 34.89% to 32.02% ( Figure 7 ).

Forest plot of studies on the influence of indoor plants on response time [ 58 , 77 , 94 ].
Heterogeneity test results of studies on the influence of indoor plants on response time.
| Model | Number of Studies | Pooled Effect Size | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
| Effect Size | Standard Error | -Value | Q-Value | df (Q) | -Value | I-Squared | ||
| Fixed | 3 | −0.252 | 0.075 | 0.001 | 51.872 | 2 | <0.001 | 96.144 |
| Random | 3 | −0.939 | 0.684 | 0.170 | ||||
3.12. Risk of Bias across Studies
Because at least three records are required for the evaluation of publication bias, only the studies investigating the effects on DBP, EEG α waves, attention, and response time were suitable for testing the risk of bias across records in the meta-analyses. All funnel plots of these studies ( Figure 8 ) revealed a symmetric funnel, confirming the absence of publication bias. Furthermore, the linear Egger’s regressions all indicated no evidence of publication bias ( p > 0.374) ( Table 20 ).

Funnel plots. ( a ) DBP; ( b ) EEG α waves; ( c ) attention; ( d ) response time.
Results of linear Egger’s regressions test.
| Egger’s Regression Test | ||
|---|---|---|
| Effect | Intercept | -Value |
| DBP | −5.892 | 0.527 |
| EEG α waves | 10.005 | 0.374 |
| attention | 7.251 | 0.656 |
| response time | −5.679 | 0.424 |
3.13. Additional Analysis
Sensitivity analysis was separately performed on records investigating the effects of indoor plants on physiological functions, including DBP and EEG α waves, and those on cognitive functions, including attention and response time, because at least three records are required. None of the pooled effect sizes changed notably when any of the studies was removed ( Figure 9 ). In summary, none of the pooled effect size values in the forest plots exceeded the 95% confidence interval of overall pooled effect size [ 95 ]. The results of the aforementioned four meta-analyses were therefore not sensitive; i.e., the results were stable and did not lead to a different conclusion if any of the included studies was deleted.

Forest plots of sensitivity analyses. ( a ) DBP [ 60 , 76 , 82 ]; ( b ) EEG α waves [ 56 , 72 , 81 ]; ( c ) attention [ 53 , 71 , 75 ]; ( d ) response time [ 58 , 77 , 94 ].
4. Discussion
The 42 records in the present systematic review provide a comprehensive perspective on the topic under investigation. Overall, the review suggests that indoor plants exerted a positive effect on objective functions in participants. Since 90.5% of the records are experiments, the above findings generally support a cause-and-effect relationship [ 49 ]. The findings on such matters, such as improved stress-reduction, increased task performance, and improved health, are in accordance with those of the previous reviews [ 31 , 36 , 40 , 45 ]. These various reviews together provide converging evidence that indoor plants are beneficial to humans, even though some reviews focused on self-reports, some on objective functions, and some did not distinguish subjective or objective responses. These findings, however, contrast with findings of no improvements in performance and productivity [ 41 ] and of no influences of indoor nature on adolescents [ 35 ]. This may be because of the differences in the measured outcomes of performance and/or functions and in the ages of the participants (cf. [ 17 ]). More studies of the effects of indoor plants on people are needed because only three systematic reviews and one meta-analysis are insufficient to draw conclusive evidence. Moreover, there are some overlapping studies between these reviews, which is not uncommon in reviews. This is also because some studies collected data on both self-reported perceptions and objectively measured responses.
4.1. Summary of Evidence
The meta-analyses covered only 16 records, consequently providing a more limited perspective than the systematic review. Nevertheless, it should be noted that synthesis findings are likely to be more reliable than those of single studies. Regarding the physiological functions, the meta-analyses further provided evidence synthesis that (1) participants exposed to indoor plants had significantly lower DBP values, which is related to excitement and arousal [ 96 ], than their counterparts exposed to no indoor plants; (2) participants exposed to indoor plants had greater EEG α waves, which is related to relaxation [ 97 ], than their counterparts, though the difference was not significant; and (3) participants exposed to indoor plants had greater EEG β waves, which is related to anxiety [ 69 ] and attention [ 98 ], than their counterparts. This difference was also not significant. It should be noted that whether the EEG wave patterns is a beneficial or an adverse function depends on the context [ 99 ]. Regarding cognitive functions, the meta-analyses further provided evidence synthesis that (1) participants exposed to indoor plants had lower attention than their counterparts, though the difference was not significant; (2) participants exposed to indoor plants had significantly higher academic achievement than their counterparts; and (3) participants exposed to indoor plants responded more quickly than their counterparts, though the difference was not significant.
Given that the pooled effect sizes of the records of DBP, EEG α waves, attention, and response time did not change notably when any record was deleted, the meta-analyses had high stability results; i.e., the removal of no study led to a different conclusion. Furthermore, the perfect homogeneity of the two studies on academic achievement provided reliable meta-analysis results (cf. [ 20 ]), although one of the included studies had low research quality, which was associated with risks of bias. Some of the results of the meta-analyses, however, were inconsistent, regardless of whether they reached significance. These included greater EEG α and β waves, and lower attention but higher academic achievement and quicker responses when exposed to indoor plants. Since there are three kinds of attention—working memory, cognitive flexibility, and attentional control—researchers should reach a consensus on which task to use to measure attention in order to have a more reliable evidence synthesis [ 19 , 20 ]. More studies of these subjects are needed.
The evidence synthesis of the relaxed physiology, as indicated by significantly lower DBP values when the participants were exposed to indoor plants than their counterparts, provided partial support to the SRT, which proposes that natural environment is helpful for recovery from stress [ 3 ], while that of the enhanced cognition, as indicated by significantly higher academic achievement when the participants exposed to indoor plants than their counterparts, provided partial support to the ART, which claims that the natural environment is beneficial to the restoration of directed attention [ 7 ]. Although different cultures may influence people’s perceptions of plants and even their functions in relation to plants (cf. [ 3 ]), there appears to be no research on these issues. Nevertheless, the evidence synthesis regarding human functions of this study seems not to be influenced by cultures. The evidence synthesis of relaxed physiology comes from the studies recruiting participants in the South Korea [ 82 ], China [ 60 ], and Taiwan [ 76 ]. Although these three studies had a significant heterogeneity (I 2 = 97.554%), the removal of any study did not change the results. Moreover, the evidence synthesis of enhanced cognition comes from the studies recruiting participants in the US [ 86 ] and Taiwan [ 66 ] but had a perfect homogeneity (I 2 = 0%). Nevertheless, more studies are needed to explore the influences of different cultures on peoples’ perceptions and functions with respect to plants.
As mentioned in the previous section concerning the risk of bias within studies, the records suffered, in general, five major risks of bias. First, noncompliance with an ITT analysis might result in unduly liberal estimate of the treatment effect [ 100 ]. Second, results obtained from unrepresentative participants might prevent observed effects from being generalizable to a larger population [ 49 ], but generalizability is improved more by many heterogeneous small experiments than by only a few large experiments [ 50 ]. Third, when outcome assessors were aware of participant allocation, outcomes might be assessed differently [ 101 ]. Fourth, statistical power not being reported might increase Type II errors: the acceptance of a null hypothesis that is actually false [ 102 ]. Fifth, the lack of appropriate randomization procedures and random allocation to groups might introduce bias [ 103 ].
Moreover, the records on the physiological and cognitive functions for the meta-analyses were susceptible to other risks of bias. Inclusion and/or exclusion criteria of participants not being reported [ 53 , 58 , 66 , 71 , 77 , 86 , 94 ] might miss the target population and/or might bias the research results [ 104 ]. Baseline measures not taken before the intervention [ 58 , 72 , 76 , 81 , 86 , 94 ] lacked a point of reference to gauge how effective the intervention is [ 49 ]. Inappropriate statistical analysis methods for study design, such as repeated-measures or within-subjects design not using repeated-measures or dependent-sample analyses [ 72 , 81 ], led to incorrect results. Not blinding participants to research questions [ 82 ] might affect their responses [ 105 ]. Lack of individual level allocation [ 58 , 81 , 86 ], in which each participant did not have an equal opportunity of being assigned to groups, might result in incomparable groups before intervention [ 50 ]. Inconsistency of intervention (within and between groups) was an issue in several studies as the intervention included more than one treatment [ 53 , 56 , 58 , 66 , 71 , 72 , 77 , 81 , 94 ], such as various plant colors.
Some studies found gender differences regarding physiological mobilization [ 57 , 62 , 69 , 83 ] and cognitive functions [ 70 , 71 ]. Such findings suggest that taking gender into consideration when investigating the effects of indoor plants is important, since males and females may have differing physiological and psychological responses (cf. [ 24 ]). Some of the records also showed different effects of plants with flowers and without flowers on physiology [ 57 , 69 , 83 ] and behavior [ 85 ]. Taking flowers and their colors and even leaf colors into account, therefore, when examining the effects of indoor plants is necessary. Moreover, most of the studies investigated the effect of only single exposure to indoor plants. Although a few studies examined the long-term effects [ 57 , 65 , 66 , 67 , 76 , 86 ], they did not scrutinize the specific effect of exposure time and/or frequency, nor did they include studies considering the influence of distance between plant and participant.
4.2. Limitations
Only journal articles were included in the review and meta-analyses, whereas grey literature was excluded. Therefore, some publication bias may have been involved [ 43 ]. There was a chance of positive [ 86 ] and/or small [ 75 , 81 , 82 ] studies being overrepresented, thus biasing the evidence synthesis. In general, studies with negative findings are less published than positive findings [ 106 ], which may give a distorted image of what is really known about a subject [ 107 , 108 ]. However, the results of DBP, EEG α waves, attention, and response time all indicated no evidence of publication bias. Furthermore, only a few records were included for the meta-analyses. Though conducting a meta-analysis with two or three studies is acceptable, it is not ideal. Since some of the 42 papers were published a long time ago, their authors could not locate the original data on means and standard deviations. Some authors could not even be reached. Additionally, five of the six meta-analyses had a very high heterogeneity (I 2 > 82%), which is associated with low reliability results [ 109 ]. This may be because of the diversity in the recruited participants, applied interventions, measured outcomes, and adopted study designs (cf. [ 109 ]). Additionally, because there were only two or three records for each of the meta-analyses, subgroup analyses, meta-regression analyses, moderating factors (gender, plant quantity, exposure duration, distance to plants, room climate, and room size), and further analyses for the risk of bias could not be conducted. Nevertheless, the results of DBP, EEG α waves, attention, and response time showed no publication bias. Moreover, because of the lack of original data on means and standard deviations or the insufficient number of studies, a meta-analysis on the effects of indoor plants on objective functions in behavior (e.g., pain tolerance and misconduct), health (sick leave, pain killer consumption, and hospitalizations), physiology (EDA, heart rate, respiration rate, and body temperature), and cognition (productivity and reaction) could not be performed. Finally, the studies included for the systematic review and those for the meta-analyses, in general, had moderate research quality (45.3% for those in the review, and 48.0% for those in the meta-analyses; [ 19 ]). Thus, high-quality research was lacking ( Table 5 and Table 6 ).
4.3. Suggestions
Future studies should recruit more people living in the equatorial area and the Global South in general and Africans in specific, preferably not college students ( Table 2 ). Background information of the participants, such as gender, age, occupation, ethnicity, health status, and number before, during, and after the research, should be provided. Study designs should use more field experiments conducted in real-world indoor environments rather than laboratories in order to improve ecological validity and still maintain sound internal validity [ 50 ]. More high-quality research is required, such as research involving experiments that follow the Consolidated Standards of Reporting Trials (CONSORT; [ 110 ]), nonrandom experiments that follow the Transparent Reporting of Evaluations with Nonrandomized Designs (TREND; [ 111 ]), and, in general, the Publication Manual of the American Psychological Association [ 112 ].
Given that indoor plants are the intervention itself, indoor plants are associated with the construct validity of the research [ 31 ] in which plant quantity, plant–participant distance, exposure time, and exposure frequency all affect the dose–response relationship (cf. [ 113 , 114 , 115 ]). Accordingly, we suggest that future studies adopt standardized measurements of the plant quantity, such as the volume percentage of the plants in an indoor environment or the visible greenness rate. The volume percentage of the plants has been used in the included studies [ 53 , 67 ], while the visible greenness rate has also been used in previous studies [ 38 ]. The visible greenness rate concerns the percentage of the plants seen by human eyes, which is an objective measurement of plants in a three-dimensional space in the field of vision [ 116 ]. Consideration of plant quantity, exposure time, frequency, and distance may assist researchers to examine rigorously how exposure to indoor plants in terms of one event or short-term period or multiple events or long-term periods affects the objective functions of individuals by means of a dose–response or exposure–outcome relationship.
In addition to the dose of and/or exposure to the indoor plants, gender difference, flower colors, flower shapes, plant colors, plant shapes [ 57 , 62 , 69 , 70 , 71 , 83 , 85 , 88 , 89 ], and cultural influences should also be considered. Furthermore, the physiology of plants, including such factors as their roots and microorganisms [ 117 , 118 ], photosynthesis, adsorption, respiration, and evapotranspiration, which are helpful for air quality and microclimate [ 40 , 47 , 119 ], may need to be considered. Similarly, researchers should also report more detailed data on room climate, room size, light condition ( Table 3 and Table 4 ), and seasonal condition, because air quality, temperature, relative humidity, light, and season also affect human comfort, performance, and health [ 120 ]. The mechanisms of and/or pathways to the effects of indoor plants on human functions also await exploration. Furthermore, indoor plants are mostly studied for their individual performances rather than as a combination. The research into the effect of plants usually focuses on the effects of single plants of different species in different conditions. Attention should further be placed on species that can cohabitate together, thus compensating each other’s needs and recreating the basic forms of symbiosis [ 121 ].
Finally, if the number of studies remains inadequate during future analyses, various aspects of human functions may be integrated into physiology (with respect to the sympathetic nervous system or parasympathetic nervous system), cognition (regarding participants’ reaction time and accuracy rate), health (in terms of illness and recovery), and behavior (either positive or negative). In that manner, the standardized mean differences (SMDs) of the function data could be adopted to conduct more rigorous meta-analyses, subgroup analyses, meta-regression analyses, and moderating factors. In contrast to self-reported measures, objective outcome measures lead to fewer reliability and validity concerns (cf. [ 122 ]) and risk of bias [ 123 ]. Nevertheless, compare and contrast of self-reported measures and objective outcome measures can provide interesting results and can be an advantage of such endeavor (c.f. [ 124 , 125 , 126 ]).
5. Conclusions
The systematic review of 42 records showed that indoor plants affect participants’ objective functions positively, particularly in terms of relaxed physiology and improved cognition. The meta-analyses further provided the evidence synthesis that indoor plants could significantly benefit participants’ SBP and academic achievement, which supported the SRT and ART. The records for the abovementioned meta-analyses, however, were limited, at only three studies for the SBP and two studies for the academic achievement. The evidence synthesis should be interpreted with caution. In brief, the systematic review concluded that, in general, people have better functions with the presence of indoor plants than the absence of indoor plants, and the meta-analyses concluded that, in specific, people have significantly lower SBP and significantly greater academic achievement when indoor plants are present than when indoor plants are not present, though with limited evidence synthesis. Since this study was the first meta-analyses of the effects of indoor plants on people’s functions, however, the findings may help the general public, environmental designers, and planners and policy makers to conduct appropriate assessments and to implement measures to improve psycho-physiological health and productivity (i.e., relaxed physiology and enhanced cognition) of habitants. The estimated productivity decrease caused by sick building syndrome, which is “a medical condition in which people in a building suffer from symptoms of illness or feeling unwell for no apparent reason” [ 127 ], in American office workers, for example, was 2%, for an annual cost of roughly 60 billion USD [ 128 ]. Furthermore, poor indoor air quality decreases workplace productivity by 10–15% [ 129 ]. The integration of plants as a building service is viable. A combination of indoor plants and ventilation technology provides enhanced efficiency and effectiveness of air purification [ 130 , 131 ]. Not only are green spaces needed in cities, but also plants are needed in buildings for people’s health and well-being. For the sake of people’s effective daily functions, indoor plants should be among the important elements of the healthy city, particularly in terms of their easy applicability and accessibility.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijerph19127454/s1 , Table S1: The full search strings; Table S2: Full-text excluded, with reason for exclusion.
Funding Statement
This research was funded by the Ministry of Science and Technology, grant number MOST 107-2410-H-167-008-MY2.
Author Contributions
Conceptualization, K.-T.H.; methodology, K.-T.H.; validation, K.-T.H.; formal analysis, K.-T.H. and L.-W.R.; investigation, K.-T.H. and L.-W.R.; resources, K.-T.H.; data curation, K.-T.H., L.-W.R. and L.-S.L.; writing—original draft preparation, K.-T.H.; writing—review and editing, K.-T.H.; visualization, L.-W.R. and L.-S.L.; supervision, K.-T.H.; project administration, K.-T.H.; funding acquisition, K.-T.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Data availability statement, conflicts of interest.
The authors declare no conflict of interest.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Plant Taxonomy: A Historical Perspective, Current Challenges, and Perspectives
- First Online: 11 December 2020
Cite this protocol

- Germinal Rouhan 3 &
- Myriam Gaudeul 3
Part of the book series: Methods in Molecular Biology ((MIMB,volume 2222))
2314 Accesses
11 Citations
Taxonomy is the science that explores, describes, names, and classifies all organisms. In this introductory chapter, we highlight the major historical steps in the elaboration of this science, which provides baseline data for all fields of biology and plays a vital role for society but is also an independent, complex, and sound hypothesis-driven scientific discipline.
In a first part, we underline that plant taxonomy is one of the earliest scientific disciplines that emerged thousands of years ago, even before the important contributions of the Greeks and Romans (e.g., Theophrastus, Pliny the Elder, and Dioscorides). In the fifteenth–sixteenth centuries, plant taxonomy benefited from the Great Navigations, the invention of the printing press, the creation of botanic gardens, and the use of the drying technique to preserve plant specimens. In parallel with the growing body of morpho-anatomical data, subsequent major steps in the history of plant taxonomy include the emergence of the concept of natural classification , the adoption of the binomial naming system (with the major role of Linnaeus) and other universal rules for the naming of plants, the formulation of the principle of subordination of characters, and the advent of the evolutionary thought. More recently, the cladistic theory (initiated by Hennig) and the rapid advances in DNA technologies allowed to infer phylogenies and to propose true natural, genealogy-based classifications.
In a second part, we put the emphasis on the challenges that plant taxonomy faces nowadays. The still very incomplete taxonomic knowledge of the worldwide flora (the so-called taxonomic impediment) is seriously hampering conservation efforts that are especially crucial as biodiversity has entered its sixth extinction crisis. It appears mainly due to insufficient funding, lack of taxonomic expertise, and lack of communication and coordination. We then review recent initiatives to overcome these limitations and to anticipate how taxonomy should and could evolve. In particular, the use of molecular data has been era-splitting for taxonomy and may allow an accelerated pace of species discovery. We examine both strengths and limitations of such techniques in comparison to morphology-based investigations, we give broad recommendations on the use of molecular tools for plant taxonomy, and we highlight the need for an integrative taxonomy based on evidence from multiple sources.
Taxonomy can justly be called the pioneering exploration of life on a little known planet. —Wilson (2004). The goal of discovering, describing, and classifying the species of our planet assuredly qualifies as big science . —Wheeler et al. (2004).
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Similar content being viewed by others

Introduction to Botanical Taxonomy

LCVP, The Leipzig catalogue of vascular plants, a new taxonomic reference list for all known vascular plants
Dobzhansky T (1973) Nothing in biology makes sense except in the light of evolution. Am Biol Teach 35:125–129
Article Google Scholar
de Carvalho MR, Bockmann FA, Amorim DS, de Vivo M, de Toledo-Piza M, Menezes NA, de Figueiredo JL, Castro RMC, Gill AC, McEachran JD, Compagno LJV, Schelly RC, Britz R, Lundberg JG, Vari RP, Nelson G (2005) Revisiting the taxonomic impediment. Science 307:353–353
Article PubMed Google Scholar
Candolle AP (1813) Théorie Élémentaire de la botanique, ou Exposition des principes de la classification naturelle et de l'art de décrire et d'étudier les végétaux
Google Scholar
Heywood VH, Watson RT (1995) Global biodiversity assessment. Cambridge University Press, Cambridge
Mayr E (1969) Principles of systematic zoology. Mcgraw-Hill, New. York
Simpson GG (1961) Principles of animal taxonomy. Columbia University Press, New York
Book Google Scholar
Tillier S (2000) Systématique - Ordonner la diversité du Vivant. Rapport sur la science et la technologie de l’Académie des sciences n°11. Éditions Tec & Doc
Small E (1989) Systematics of biological systematics (or taxonomy of taxonomy). Taxon 38:335–356
Sprague JL, Lanjouw J, Andreas CH (1948) Minutes of the Utrecht conference. Chronica Botanica 12(1/2):12
Morton CV (1957) The misuse of the term taxon. Taxon 6(5):155
Raven PH (2004) Taxonomy: where are we now? Philos Trans R Soc Lond B Biol Sci 359:729–730
Article PubMed PubMed Central Google Scholar
Pavord A (2005) The naming of names: the search for order in the world of plants. Bloomsbury, New York
Funk VA, Hoch PC, Prather LA, Wagner WL (2005) The importance of vouchers. Taxon 54:127–129
Knapp S (2012) What’s in a name? A history of taxonomy. http://www.nhm.ac.uk/nature-online/science-of-natural-history/taxonomy-systematics/history-taxonomy . Accessed Jan 2012
Griffing LR (2011) Who invented the dichotomous key? Richard Waller’s watercolors of the herbs of Britain. Am J Bot 98:1911–1923
Linnaeus C (1753) Species Plantarum. Stockholm
Linnaeus C (1758) Systema naturae, 10th edn. Stockholm
Candolle AP (1867) Lois de la nomenclature botanique adoptées par le Congrès international de botanique: tenu à Paris en août 1867. H. Georg, Geneva
Jussieu AL (1789) Genera plantarum. Herissant, Paris
Philippe H, Lecointre G, VanLe HL, LeGuyader H (1996) A critical study of homoplasy in molecular data with the use of a morphologically based cladogram, and its consequences for character weighting. Mol Biol Evol 13:1174–1186
Lamarck J-BPAM (1809) Philosophie zoologique. Dentu, Paris
Darwin C (1859) On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray, London
Haeckel E (1866) Generelle Morphologie der Organismen. Reimer, Berlin
Dayrat B (2003) The roots of phylogeny: how did Haeckel build his trees? Syst Biol 52:515–527
Davis PH, Heywood PH (1963) Principles of angiosperm taxonomy. Oliver and Boyd, Edinburgh/London
Sneath PHA, Sokal RR (1963) Principles of numerical taxonomy, 7th edn. W. H. Freeman, San Francisco
Hennig W (1950) Grundzüge einer Theorie der phylogenetischen Systematik. Deutscher Zentralverlag, Berlin
Hennig W (1966) Phylogenetic systematics (tr. D. Davis and R. Zangerl), University of Illinois Press, Urbana
Godfray HCJ, Knapp S (2004) Taxonomy for the twenty-first century - introduction. Philos Trans R Soc Lond B Biol Sci 359:559–569
Article CAS PubMed PubMed Central Google Scholar
Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H (1986) Specific enzymatic amplification of DNA Invitro - the polymerase chain-reaction. Cold Spring Harb Symp Quant Biol 51:263–273
Article CAS PubMed Google Scholar
Bremer K, Chase MW, Stevens PF, Anderberg AA, Backlund A, Bremer B, Briggs BG, Endress PK, Fay MF, Goldblatt P, Gustafsson MHG, Hoot SB, Judd WS, Kallersjo M, Kellogg EA, Kron KA, Les DH, Morton CM, Nickrent DL, Olmstead RG, Price RA, Quinn CJ, Rodman JE, Rudall PJ, Savolainen V, Soltis DE, Soltis PS, Sytsma KJ, Thulin M, Grp AP (1998) An ordinal classification for the families of flowering plants. Ann Mo Bot Gard 85:531–553
Bremer B, Bremer K, Chase MW, Reveal JL, Soltis DE, Soltis PS, Stevens PF, Anderberg AA, Fay MF, Goldblatt P, Judd WS, Kallersjo M, Karehed J, Kron KA, Lundberg J, Nickrent DL, Olmstead RG, Oxelman B, Pires JC, Rodman JE, Rudall PJ, Savolainen V, Sytsma KJ, van der Bank M, Wurdack K, Xiang JQY, Zmarzty S, Grp AP (2003) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141:399–436
Bremer B, Bremer K, Chase MW, Fay MF, Reveal JL, Soltis DE, Soltis PS, Stevens PF, Anderberg AA, Moore MJ, Olmstead RG, Rudall PJ, Sytsma KJ, Tank DC, Wurdack K, Xiang JQY, Zmarzty S, Grp AP (2009) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161:105–121
The Angiosperm phylogeny group (2016) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181:1–20. https://doi.org/10.1111/boj.12385
PPGI (2016) A community-derived classification for extant lycophytes and ferns. J Syst Evol 54:563–603. https://doi.org/10.1111/jse.12229
Pryer KM, Schneider H, Smith AR, Cranfill R, Wolf PG, Hunt JS (2001) Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409:618–621. https://doi.org/10.1038/35054555
Bessey CE (1915) The phylogenetic taxonomy of flowering plants. Ann Mo Bot Gard 2:109–164
Cronquist A (1981) An integrated system of classification of flowering plants. Columbia University Press, New York
Stebbins GL (1974) Flowering plants: evolution above the species level. Belknap press, Cambridge
Takhtajan A (1997) Diversity and classification of flowering plants. Columbia University Press, New York
Thorne RF (1976) A phylogenetic classification of the Angiospermae. Evol Biol 9:35–106
Soltis DE, Soltis PS, Endress PK, Chase MW (2005) Phylogeny and evolution of angiosperms. Sinauer associates, Sunderland
Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 270:313–321
Article CAS Google Scholar
May RM (2004) Tomorrow's taxonomy: collecting new species in the field will remain the rate-limiting step. Philos Trans R Soc Lond B Biol Sci 359:733–734
May RM (2011) Why worry about how many species and their loss? PLoS Biol 9:e1001130
Paton AJ, Brummitt N, Govaerts R, Harman K, Hinchcliffe S, Allkin B, Lughadha EN (2008) Towards target 1 of the global strategy for plant conservation: a working list of all known plant species - progress and prospects. Taxon 57:602–611
Wilson EO (2003) The encyclopedia of life. Trends Ecol Evol 18:77–80
Wilson EO (2004) Taxonomy as a fundamental discipline. Philos Trans R Soc Lond B Biol Sci 359:739–739
Candolle AP (1824-1873) Prodromus systematis naturalis regni vegetabilis. Sumptibus Sociorum Treuttel et Würtz, Parisii
International Plant Names Index (2012) Published on the Internet http://www.ipni.org . Accessed July 2019
Lughadha EN, Govaerts R, Belyaeva I, Black N, Lindon H, Allkin R, Magill RE, Nicolson N (2016) Counting counts: revised estimates of numbers of accepted 407 species of flowering plants, seed plants, vascular plants and land plants with a review 408 of other recent estimates. Phytotaxa 272:82–88
Scotland RW, Wortley AH (2003) How many species of seed plants are there? Taxon 52:101–104
The Plant List (2019) Version 1. Published on the Internet http://www.theplantlist.org/ . Accessed July 2019
Wortley AH, Scotland RW (2004) Synonymy, sampling and seed plant numbers. Taxon 53:478–480
Mallet J, Willmott K (2003) Taxonomy: renaissance or tower of babel? Trends Ecol Evol 18:57–59
Isaac NJB, Mallet J, Mace GM (2004) Taxonomic inflation: its influence on macroecology and conservation. Trends Ecol Evol 19:464–469
Meiri S, Mace GM (2007) New taxonomy and the origin of species. PLoS Biol 5:1385–1386
Pillon Y, Chase MW (2007) Taxonomic exaggeration and its effects on orchid conservation. Conserv Biol 21:263–265
Crane PR (2004) Documenting plant diversity: unfinished business. Philos Trans R Soc Lond B Biol Sci 359:735–737
Joppa LN, Roberts DL, Pimm SL (2011) How many species of flowering plants are there? Proc R Soc B Biol Sci 278:554–559
Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B (2011) How many species are there on earth and in the ocean? PLoS Biol 9:e1001127
Bisby FA, Roskov YR, Orrell TM, Nicolson D, Paglinawan LE et al (2010) Species 2000 & ITIS Catalogue of Life: 2010 Annual Checklist. Digital resource at http://www.catalogueoflife.org/annual-checklist/2010 . Species 2000, Reading, UK
Caldecott JO, Jenkins MD, Johnson TH, Groombridge B (1996) Priorities for conserving global species richness and endemism. Biodivers Conserv 5:699–727
Joppa LN, Roberts DL, Myers N, Pimm SL (2011) Biodiversity hotspots house most undiscovered plant species. Proc Natl Acad Sci U S A 108:13171–13176
Callmander MW, Schatz GE, Lowry PP (2005) IUCN red list assessment and the global strategy for plant conservation: taxonomists must act now. Taxon 54:1047–1050
Godfray HCJ (2002) Challenges for taxonomy - the discipline will have to reinvent itself if it is to survive and flourish. Nature 417:17–19
Funk VA (2006) Floras: a model for biodiversity studies or a thing of the past? Taxon 55:581–588
Wheeler QD, Raven PH, Wilson EO (2004) Taxonomy: impediment or expedient? Science 303:285–285
Wheeler QD, Knapp S et al (2012) Mapping the biosphere: exploring species to understand the origin, organization and sustainability of biodiversity. Syst Biodivers 10(1):1–20
Ebach MC, Valdecasas AG, Wheeler QD (2011) Impediments to taxonomy and users of taxonomy: accessibility and impact evaluation. Cladistics 27:550–557
Ronquist F, Gardenfors U (2003) Taxonomy and biodiversity inventories: time to deliver. Trends Ecol Evol 18:269–270
Joly CA (2006) Taxonomy: programmes developing in the south too. Nature 440:24–24
Schatz GE, Lowry PP, Ramisamihantanirina A (1998) Takhtajania perrieri rediscovered. Nature 391:133–134
Jones WG, Hill KD, Allen JM (1995) Wollemia nobilis , a new living Australian genus and species in the Araucariaceae. Telopea 6:173–176
Mabberley DJ (2009) Exploring Terra Incognita. Science 324:472–472
Thulin M (2007) Acacia fumosa sp nov (Fabaceae) from eastern Ethiopia. Nord J Bot 25:272–274
Dransfield J, Rakotoarinivo M, Baker WJ, Bayton RP, Fisher JB, Horn JW, Leroy B, Metz X (2008) A new coryphoid palm genus from Madagascar. Bot J Linn Soc 156:79–91
Agnarsson I, Kuntner M (2007) Taxonomy in a changing world: seeking solutions for a science in crisis. Syst Biol 56:531–539
Crisci JV (2006) One-dimensional systematist: perils in a time of steady progress. Syst Bot 31:217–221
Joppa LN, Roberts DL, Pimm SL (2011) The population ecology and social behaviour of taxonomists. Trends Ecol Evol 26:551–553
Rodman JE, Cody JH (2003) The taxonomic impediment overcome: NSF's partnerships for enhancing expertise in taxonomy (PEET) as a model. Syst Biol 52:428–435
Bebber DP et al (2012) Big hitting collectors make massive and disproportionate contribution to the discovery of plant species. Proc R Soc Lond B Biol Sci 279(1736):2269–2274
Thiers B (2019) Index herbarium: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/ih/
Bebber DP, Carine MA, Wood JRI, Wortley AH, Harris DJ, Prance GT, Davidse G, Paige J, Pennington TD, Robson NKB, Scotland RW (2010) Herbaria are a major frontier for species discovery. Proc Natl Acad Sci U S A 107:22169–22171
Fontaine B, Perrard A, Bouchet P (2012) 21 years of shelf life between discovery and description of new species. Curr Biol 22(22):R943–R944. https://doi.org/10.1016/j.cub.2012.10.029
Le Bras G, Pignal M, Jeanson ML, Muller S, Aupic C, Carré B, Flament G, Gaudeul M, Gonçalves C, Invernón VR, Jabbour F, Lerat E, Lowry PP, Offroy B, Pérez Pimparé E, Poncy O, Rouhan G, Haevermans T (2017) The French Muséum national d’histoire naturelle vascular plant herbarium collection dataset. Scientific Data 4:170016
Godfray HCJ, Clark BR, Kitching IJ, Mayo SJ, Scoble MJ (2007) The web and the structure of taxonomy. Syst Biol 56:943–955
Knapp S, McNeill J, Turland NJ (2011) Changes to publication requirements made at the XVIII International Botanical Congress in Melbourne - what does e-publication mean for you? PhytoKeys (6):5–11
Nicolson N, Challis K, Tucker A, Knapp S (2017) Impact of e-publication changes in the International Code of Nomenclature for algae, fungi and plants (Melbourne Code, 2012) - did we need to “run for our lives”? BMC Evol Biol 17:116. https://doi.org/10.1186/s12862-017-0961
Hebert PDN, Gregory TR (2005) The promise of DNA barcoding for taxonomy. Syst Biol 54:852–859
Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R (2005) Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philos Trans R Soc B Biol Sci 360:1805–1811
Wiens JJ (2007) Species delimitation: new approaches for discovering diversity. Syst Biol 56:875–878
Pannell JR (2009) Mating-system evolution: succeeding by celibacy. Curr Biol 19:R983–R985
Hood ME, Antonovics J (2003) Plant species descriptions show signs of disease. Proc R Soc Lond B Biol Sci 270:S156–S158
Duminil J, Di Michele M (2009) Plant species delimitation: a comparison of morphological and molecular markers. Plant Biosystems 143:528–542
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155
Grundt HH, Kjolner S, Borgen L, Rieseberg LH, Brochmann C (2006) High biological species diversity in the arctic flora. Proc Natl Acad Sci U S A 103:972–975
Pillon Y, Hopkins HCF, Munzinger J, Amir H, Chase MW (2009) Cryptic species, gene recombination and hybridization in the genus Spiraeanthemum (Cunoniaceae) from New Caledonia. Bot J Linn Soc 161:137–152
Dulvy NK, Reynolds JD (2009) BIODIVERSITY skates on thin ice. Nature 462:417–417
Robertson A, Newton AC, Ennos RA (2004) Multiple hybrid origins, genetic diversity and population genetic structure of two endemic Sorbus taxa on the Isle of Arran, Scotland. Mol Ecol 13:123–134
Squirrell J, Hollingsworth PM, Bateman RM, Tebbitt MC, Hollingsworth ML (2002) Taxonomic complexity and breeding system transitions: conservation genetics of the Epipactis leptochila complex (Orchidaceae). Mol Ecol 11:1957–1964
van Dijk PJ (2003) Ecological and evolutionary opportunities of apomixis: insights from Taraxacum and Chondrilla. Philos Trans R Soc Lond Ser B Biol Sci 358:1113–1121
Ennos RA, French GC, Hollingsworth PM (2005) Conserving taxonomic complexity. Trends Ecol Evol 20:164–168
Ennos RA, Whitlock R, Fay MF, Jones B, Neaves LE, Payne R, Taylor I, De Vere N, Hollingsworth PM (2012) Process-based species action plans: an approach to conserve contemporary evolutionary processes that sustain diversity in taxonomically complex groups. Bot J Linn Soc 168:194–203
Li FW, Tan BC, Buchbender V, Moran RC, Rouhan G, Wang CN, Quandt D (2009) Identifying a mysterious aquatic fern gametophyte. Plant Syst Evol 281:77–86
Van Deynze A, Stoffel K (2006) High-throughput DNA extraction from seeds. Seed Sci Technol 34:741–745
Asif MJ, Cannon CH (2005) DNA extraction from processed wood: a case study for the identification of an endangered timber species (Gonystylus bancanus). Plant Mol Biol Report 23:185–192
Colpaert N, Cavers S, Bandou E, Caron H, Gheysen G, Lowe AJ (2005) Sampling tissue for DNA analysis of trees: trunk cambium as an alternative to canopy leaves. Silvae Genetica 54:265–269
Rachmayanti Y, Leinemann L, Gailing O, Finkeldey R (2006) Extraction, amplification and characterization of wood DNA from Dipterocarpaceae. Plant Mol Biol Report 24:45–55
Tibbits JFG, McManus LJ, Spokevicius AV, Bossinger G (2006) A rapid method for tissue collection and high-throughput isolation of genomic DNA from mature trees. Plant Mol Biol Report 24:81–91
Novaes RML, Rodrigues JG, Lovato MB (2009) An efficient protocol for tissue sampling and DNA isolation from the stem bark of Leguminosae trees. Genet Mol Res 8:86–96
Deguilloux MF, Pemonge MH, Petit RJ (2002) Novel perspectives in wood certification and forensics: dry wood as a source of DNA. Proc R Soc B Biol Sci 269:1039–1046
Hiiesalu I, Opik M, Metsis M, Lilje L, Davison J, Vasar M, Moora M, Zobel M, Wilson SD, Partel M (2012) Plant species richness belowground: higher richness and new patterns revealed by next-generation sequencing. Mol Ecol 21:2004–2016
Kesanakurti PR, Fazekas AJ, Burgess KS, Percy DM, Newmaster SG, Graham SW, Barrett SCH, Hajibabaei M, Husband BC (2011) Spatial patterns of plant diversity below-ground as revealed by DNA barcoding. Mol Ecol 20:1289–1302
Dunn CP (2003) Keeping taxonomy based in morphology. Trends Ecol Evol 18:270–271
Santos LM, Faria LRR (2011) The taxonomy’s new clothes: a little more about the DNA-based taxonomy. Zootaxa 3025:66–68
Schaefer H, Carine MA, Rumsey FJ (2011) From European priority species to invasive weed: Marsilea azorica (Marsileaceae) is a misidentified alien. Syst Bot 36:845–853
Launert GOE, Paiva JAR (1983) Iconographia selecta florae Azoricae. Coimbra 2:159
Lipscomb D, Platnick N, Wheeler Q (2003) The intellectual content of taxonomy: a comment on DNA taxonomy. Trends Ecol Evol 18:65–66
Sites JW, Marshall JC (2004) Operational criteria for delimiting species. Annu Rev Ecol Evol Syst 35:199–227
Stace CA (2005) Plant taxonomy and biosystematics - does DNA provide all the answers? Taxon 54:999–1007
Linder CR, Rieseberg LH (2004) Reconstructing patterns of reticulate evolution UN plants. Am J Bot 91:1700–1708
Vriesendorp B, Bakker FT (2005) Reconstructing patterns of reticulate evolution in angiosperms: what can we do? Taxon 54:593–604
Egan AN, Schlueter J, Spooner DM (2012) Applications of next-generation sequencing in plant biology. Am J Bot 99:175–185
Harrison N, Kidner CA (2011) Next-generation sequencing and systematics: what can a billion base pairs of DNA sequence data do for you? Taxon 60:1552–1566
Straub SCK, Parks M, Weitemier K, Fishbein M, Cronn RC, Liston A (2012) Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot 99:349–364
Bakker FT (2015) DNA sequences from plant herbarium tissue (Chapter 8). In: Hörandl E, Appelhans MS (eds) Next-generation sequencing in plant systematics. International Association for Plant Taxonomy (IAPT). https://doi.org/10.14630/000009
Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM (2001) Rapid diversification of a species-rich genus of neotropical rain forest trees. Science 293:2242–2245
Baldwin BG, Sanderson MJ (1998) Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc Natl Acad Sci U S A 95:9402–9406
Wang AL, Yang MH, Liu JQ (2005) Molecular phylogeny, recent radiation and evolution of gross morphology of the rhubarb genus Rheum (Polygonaceae) inferred from chloroplast DNA trnL-F sequences. Ann Bot 96:489–498
Hodges SA, Arnold ML (1994) Columbines - a geographically widespread species flock. Proc Natl Acad Sci U S A 91:5129–5132
Linder HP (2008) Plant species radiations: where, when, why? Philos Trans R Soc B Biol Sci 363:3097–3105
Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP (2003) A plea for DNA taxonomy. Trends Ecol Evol 18:70–74
Hey J, Pinho C (2012) Population genetics and objectivity in species diagnosis. Evolution 66:1413–1429
Knowles L, Carstens B (2007) Delimiting species without monophyletic gene trees. Syst Biol 56:887–895
Staats M, Cuenca A, Richardson JE, Vrielink-van Ginkel R, Petersen G, Seberg O, Bakker FT (2011) DNA damage in plant herbarium tissue. PLoS One 6:e28448
Seberg O, Humphries CJ, Knapp S, Stevenson DW, Petersen G, Scharff N, Andersen NM (2003) Shortcuts in systematics? A commentary on DNA-based taxonomy. Trends Ecol Evol 18:63–65
Lister DL, Bower MA, Howe CJ, Jones MK (2008) Extraction and amplification of nuclear DNA from herbarium specimens of emmer wheat: a method for assessing DNA preservation by maximum amplicon length recovery. Taxon 57:254–258
Wandeler P, Hoeck PEA, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends Ecol Evol 22:634–642
Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A (2007) Genetic variation in time and space: the use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris. Conserv Genet 8:629–639
Erkens RHJ, Cross H, Maas JW, Hoenselaar K, Chatrou LW (2008) Assessment of age and greenness of herbarium specimens as predictors for successful extraction and amplification of DNA. Blumea 53:407–428
Drabkova L, Kirschner J, Vlcek C (2002) Comparison of seven DNA extraction and amplification protocols in historical herbarium specimens of Juncaceae. Plant Mol Biol Report 20:161–175
Jankowiak K, Buczkowska K, Szweykowska-Kulinska Z (2005) Successful extraction of DNA from 100-year-old herbarium specimens of the liverwort Bazzania trilobata. Taxon 54:335–336
Korpelainen H, Pietilainen M (2008) Effort to reconstruct past population history in the fern Blechnum spicant. J Plant Res 121:293–298
Savolainen V, Cuenoud P, Spichiger R, Martinez MDP, Crevecoeur M, Manen JF (1995) The use of herbarium specimens in DNA Phylogenetics - evaluation and improvement. Plant Syst Evol 197:87–98
Ribeiro RA, Lovato MB (2007) Comparative analysis of different DNA extraction protocols in fresh and herbarium specimens of the genus Dalbergia. Genet Mol Res 6:173–187
CAS PubMed Google Scholar
Andreasen K, Manktelow M, Razafimandimbison SG (2009) Successful DNA amplification of a more than 200-year-old herbarium specimen: recovering genetic material from the Linnaean era. Taxon 58:959–962
Ames M, Spooner DM (2008) DNA from herbarium specimens settles a controversy about origins of the European potato. Am J Bot 95:252–257
Walters C, Reilley AA, Reeves PA, Baszczak J, Richards CM (2006) The utility of aged seeds in DNA banks. Seed Sci Res 16:169–178
Alves RJV, Machado MD (2007) Is classical taxonomy obsolete? Taxon 56:287–288
DeSalle R, Egan MG, Siddall M (2005) The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos Trans R Soc B Biol Sci 360:1905–1916
DeSalle R (2006) Species discovery versus species identification in DNA barcoding efforts: response to Rubinoff. Conserv Biol 20:1545–1547
Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH (2010) Integrative taxonomy: a multisource approach to exploring biodiversity. Annu Rev Entomol 55:421–438
Dayrat B (2005) Towards integrative taxonomy. Biol J Linn Soc 85:407–415
Wheeler QD (2005) Losing the plot: DNA “barcodes” and taxonomy. Cladistics 21:405–407
Corney DPA, Clark JY, Tang HT, Wilkin P (2012) Automatic extraction of leaf characters from herbarium specimens. Taxon 61(1):231–244
Raxworthy CJ, Ingram CM, Rabibisoa N, Pearson RG (2007) Applications of ecological niche modeling for species delimitation: a review and empirical evaluation using day geckos (Phelsuma) from Madagascar. Syst Biol 56:907–923
Rissler LJ, Apodaca JJ (2007) Adding more ecology into species delimitation: ecological niche models and phylogeography help define cryptic species in the black salamander (Aneides flavipunctatus). Syst Biol 56:924–942
Wiens JJ, Graham CH (2005) Niche conservatism: integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Syst 36:519–539
Brautigam A, Gowik U (2010) What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol 12:831–841
Cronn R, Knaus BJ, Liston A, Maughan PJ, Parks M, Syring JV, Udall J (2012) Targeted enrichment strategies for next-generation plant biology. Am J Bot 99:291–311
Rodriguez-Fernandez JI, De Carvalho CJB, Pasquini C, De Lima KMG, Moura MO, Arizaga GGC (2011) Barcoding without DNA? Species identification using near infrared spectroscopy. Zootaxa 46–54
Munck L, Jespersen BM, Rinnan A, Seefeldt HF, Engelsen MM, Norgaard L, Engelsen SB (2010) A physiochemical theory on the applicability of soft mathematical models-experimentally interpreted. J Chemom 24:481–495
Cruickshank RH, Munck L (2011) It’s barcoding Jim, but not as we know it. Zootaxa 2933:55–56
Andres-Sanchez S, Rico E, Herrero A, Santos-Vicente M, Martinez-Ortega MM (2009) Combining traditional morphometrics and molecular markers in cryptic taxa: towards an updated integrative taxonomic treatment for Veronica subgenus Pentasepalae (Plantaginaceae sensu APG II) in the western Mediterranean. Bot J Linn Soc 159:68–87
Schlick-Steiner BC, Seifert B, Stauffer C, Christian E, Crozier RH, Steiner FM (2007) Without morphology, cryptic species stay in taxonomic crypsis following discovery. Trends Ecol Evol 22:391–392
Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E (2005) Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc B Biol Sci 360:1935–1943
Markmann M, Tautz D (2005) Reverse taxonomy: an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Philos Trans R Soc B Biol Sci 360:1917–1924
Pleijel F, Jondelius U, Norlinder E, Nygren A, Oxelman B, Schander C, Sundberg P, Thollesson M (2008) Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Mol Phylogenet Evol 48:369–371
Puillandre N, Bouchet P, Boisselier-Dubayle MC, Brisset J, Buge B, Castelin M, Chagnoux S, Christophe T, Corbari L, Lambourdiere J, Lozouet P, Marani G, Rivasseau A, Silva N, Terryn Y, Tillier S, Utge J, Samadi S (2012) New taxonomy and old collections: integrating DNA barcoding into the collection curation process. Mol Ecol Resour 12:396–402
Gemeinholzer B, Bachmann K (2005) Examining morphological and molecular diagnostic character states of Cichorium intybus L. (Asteraceae) and C-spinosum L. Plant Syst Evol 253:105–123
Bacon CD, McKenna MJ, Simmons MP, Wagner WL (2012) Evaluating multiple criteria for species delimitation: an empirical example using Hawaiian palms (Arecaceae: Pritchardia). BMC Evol Biol 12:23
Barrett CF, Freudenstein JV (2011) An integrative approach to delimiting species in a rare but widespread mycoheterotrophic orchid. Mol Ecol 20:2771–2786
Koffi KG, Heuertz M, Doumenge C, Onana JM, Gavory F, Hardy OJ (2010) A combined analysis of morphological traits, chloroplast and nuclear DNA sequences within Santiria trimera (Burseraceae) suggests several species following the biological species concept. Plant Ecol Evol 143:160–169
Ley AC, Hardy OJ (2010) Species delimitation in the central African herbs Haumania (Marantaceae) using georeferenced nuclear and chloroplastic DNA sequences. Mol Phylogenet Evol 57:859–867
Meudt HM, Lockhart PJ, Bryant D (2009) Species delimitation and phylogeny of a New Zealand plant species radiation. BMC Evol Biol 9:111
Article PubMed PubMed Central CAS Google Scholar
Schmidt-Lebuhn AN (2007) Using amplified fragment length polymorphism (AFLP) to unravel species relationships and delimitations in Minthostachys (Labiatae). Bot J Linn Soc 153:9–19
Zeng YF, Liao WJ, Petit RJ, Zhang DY (2010) Exploring species limits in two closely related Chinese oaks. PLoS One 5:e15529
Rieseberg LH, Troy TE, Baack EJ (2006) The nature of plant species. Nature 440:524–527
de Queiroz K (2005) Ernst Mayr and the modern concept of species. Proc Natl Acad Sci U S A 102:6600–6607
de Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56(6):879–886
Wright S (1940) The statistical consequences of Mendelian heredity in relation to speciation. In: Huxley J (ed) The new systematics. Oxford university press, London, pp 161–183
Mayr E (1942) Systematics and the origin of species. Columbia university press, New York
Dobzhansky T (1950) Mendelian populations and their evolution. Am Nat 84:401–418
Poulton EB (1904) What is a species? Proc Entomol Soc Lond 1903:lxxviicxvi
Dobzhansky T (1970) Genetics of the evolutionary process. Columbia University Press, New York
Sokal RR, Crovello TJ (1970) The biological species concept: a critical evaluation. Am Nat 104:107–123
Van Valen L (1976) Ecological species, multispecies, and oaks. Taxon 25:233–239
Simpson GG (1951) The species concept. Evolution 5:285–298
Wiley EO (1978) The evolutionary species concept reconsidered. Syst Zool 21:17–26
Cracraft J (1989) Speciation and its ontology: the empirical consequences of alternative species concepts for understanding patterns and processes of differentiation. In: Otte D, Endler JA (eds) Speciation and its consequences. Sinauer Associates, Sunderland, pp 28–59
Rosen DE (1979) Fishes from the uplands and intermontane basins of Guatemala: revisionary studies and comparative geography. Bull Am Mus Nat Hist 162:267–376
Donoghue MJ (1985) A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist 88:172–181
Mishler BD (1985) The morphological, developmental, and phylogenetic basis of species concepts in bryophytes. Bryologist 88:207–214
Baum DA, Shaw KL (1995) Genealogical perspectives on the species problem. In: Hoch PC, Stephenson AG (eds) Experimental and molecular approaches to plant biosystematics. Missouri Botanical Garden, St. Louis, pp 289–303
Mallet J (1995) A species definition for the modern synthesis. Trends Ecol Evol 10:294–299
Templeton AR (1998) Species and speciation: geography, population structure, ecology, and gene trees. In: Howard DJ, Berlocher SH (eds) Endless forms: species and speciation. Oxford university press, New York, pp 32–43
Sites JW, Marshall JC (2003) Delimiting species: a renaissance issue in systematic biology. Trends Ecol Evol 18(9):462–470
Download references
Author information
Authors and affiliations.
Institut de Systématique, Evolution, Biodiversité (ISYEB), Muséum national d’Histoire naturelle, Sorbonne Université, Ecole Pratique des Hautes Etudes, Université des Antilles, CNRS, Paris, France
Germinal Rouhan & Myriam Gaudeul
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Germinal Rouhan .
Editor information
Editors and affiliations.
UMR PVBMT, Universite de la Reunion, St Pierre, Réunion, France
Pascale Besse
Rights and permissions
Reprints and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Rouhan, G., Gaudeul, M. (2021). Plant Taxonomy: A Historical Perspective, Current Challenges, and Perspectives. In: Besse, P. (eds) Molecular Plant Taxonomy. Methods in Molecular Biology, vol 2222. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0997-2_1
Download citation
DOI : https://doi.org/10.1007/978-1-0716-0997-2_1
Published : 11 December 2020
Publisher Name : Humana, New York, NY
Print ISBN : 978-1-0716-0996-5
Online ISBN : 978-1-0716-0997-2
eBook Packages : Springer Protocols
- Publish with us
Policies and ethics
- Find a journal
- Track your research

The Science of Plants
Tom Michaels, Saint Paul, MN
Emily Hoover, Saint Paul, MN
Laura Irish, Saint Paul, MN
Copyright Year: 2022
ISBN 13: 9781946135872
Publisher: University of Minnesota Libraries Publishing
Language: English
Formats Available
Conditions of use.
Learn more about reviews.
Reviewed by Paula Mejia Velasquez, Adjunct Professor, Leeward Community College on 12/8/22
This book is a great resource for an introductory-level class on Horticulture or botany, covering most of the topics usually addressed in a class at the undergrad level. Some topics are explained in more detail than others, but all the topics... read more
Comprehensiveness rating: 4 see less
This book is a great resource for an introductory-level class on Horticulture or botany, covering most of the topics usually addressed in a class at the undergrad level. Some topics are explained in more detail than others, but all the topics presented in the book are well explained. An important topic that is not included in the book is ecology, explaining different ways that plants interact with other organisms (e.g. animals, plants, fungi).
Content Accuracy rating: 5
The content of the book appears accurate, and the topics are presented in a neutral, not biased way.
Relevance/Longevity rating: 5
The topics covered in the book are relevant for an introductory plant science class at an undergrad level. In terms of longevity, most of the material should still be relevant in the long term, but other topics, like taxonomy, would probably need to be updated every so often.
Clarity rating: 5
The book is easy to follow and read, at a level that is accessible and understandable for undergrad students. A list of terms or glossary is included in each chapter and at the end of the book, this helps students that need more explanation on a term.
Consistency rating: 5
The book is consistent from beginning to end, presenting a similar writing style and format.
Modularity rating: 5
The organization of the chapters and the subunits is clear and consistent. Individual chapters or subunits can be found easily on the chapter outline, making it easy to alter the order of the book content based on teaching preferences.
Organization/Structure/Flow rating: 2
Each chapter consists of a brief introduction, learning objectives, the topic per se, and a glossary. This structure is similar to most textbooks and I think it works great. However, I had a hard time with the order that the topics are presented in this book. In my opinion, the organization of the chapters could follow a different order to improve the flow of the book. There are several concepts/topics in the book that are grouped with concepts/topics that do not seem to be closely related, and that maybe would fit better under other more related chapters. For example, flower morphology is grouped in a chapter with meristems, plant taxonomy is grouped with seed germination, and plant growth is grouped with inflorescences. It would probably make more sense to group flower morphology with inflorescences, and meristems with plant growth. In a specific example, seed germination is presented in Chapter 2 as section 2.2, but a more comprehensive chapter on seeds is included in Chapter 9. I think the seed germination section would be a better fit for Chapter 9, and leave Chapter 2 as a taxonomic chapter.
Interface rating: 4
The textbook is available in different formats, including pdf, word, xml, ebook, and online. I explored the online and pdf versions and found them easy to navigate. The online version includes short videos that expand on some topics, as well as embedded h5p interactive activities to test comprehension and increase student engagement. The pdf version on the other hand does not have the videos or interactive activities embedded, instead, it provides links to them. However, some of the links provided do not work (e.g. links on pages 12 -13). I would recommend using the online version of the book.
Grammatical Errors rating: 5
The text contains no significant amount of grammatical errors.
Cultural Relevance rating: 5
I did not find the book to be culturally insensitive or offensive. It is mainly focused on plants found in northern temperate areas. Including some tropical examples could make this book more attractive to a wider audience.
This book does a great job of covering and explaining basic plant and horticultural science concepts that are typically included in an introductory-level botany or horticulture class. I really liked that it includes videos, a glossary, interactive activities (i.e. h5p), and other supplementary materials (i.e. review questions and Quizlet flashcards), as I think they are great tools to complement the content of the book and engage students. The book is easy to read, each chapter includes specific learning outcomes, a chapter outline, a summary, and review questions. I disagree with the order the topics are presented, but each instructor could easily address this by assigning the chapters in a different order.
Table of Contents
Chapter 1: Plants in our Lives
1.1 What is horticulture?
1.2 Science and Experimentation
1.3 Plant Parts we Eat
Chapter 1: Terms
Chapter 2: Taxonomy and Seed Germination
2.1 Plant Taxonomy
2.2 Introduction to Seed Germination
Chapter 2: Terms
Chapter 3: How Plants Grow, Part 1
Chapter 3: Terms
Chapter 4: How Plants Grow, Part 2
4.1 Growth Patterns and Inflorescences
4.2 Plant Hormones
Chapter 4: Terms
Chapter 5: Inside Plants
5.1 Inside Leaves
5.2 Inside Stems
5.3 Inside Roots
Chapter 5: Terms
Chapter 6: Cells, Tissues, and Woody Growth
6.1 Plant Cells and Tissues
6.2 Woody Growth
Chapter 6: Terms
Chapter 7: Meristems and Flowers
7.1 Meristem Morphology
7.2 Flower Morphology
Chapter 7: Terms
Chapter 8: Fruit
8.1 Fruit Morphology
Chapter 8: Terms
Chapter 9: Seeds
9.1 Seed Morphology
9.2 Seed Physiology
Chapter 9: Terms
Chapter 10: Grafting
10.1 Grafts and Wounds
10.2 Unique Storage Organs
Chapter 10: Terms
Chapter 11: Water and Light
11.1 Plants and Water
11.2 Light and Photosynthesis
Chapter 11: Terms
Chapter 12: Soils, Fertility, and Plant Growth
12.1 Soils, Fertility, and Plant Growth
Chapter 12: Terms
Chapter 13: Sexual Reproduction
13.2 Mitosis
13.3 Meiosis
Chapter 13: Terms
Chapter 14: Variation and Plant Breeding
14.1 Gametogenesis
14.2 Inheritance of Big Differences
14.3 Linkage and Inheritance of Small Differences
14.4 Plant Breeding
Chapter 14: Terms
Chapter 15: Invasive plants and GMOs
15.1 Invasive plants
Chapter 15: Terms
Glossary of Terms
Ancillary Material
About the book.
An approachable guide to the fundamentals of plant science. Created for horticulture students, gardeners, science teachers, and anyone interested in understanding plants and how they grow. This is the required text for HORT 1001/6001 Plant Propagation at the University of Minnesota Department of Horticultural Science.
About the Contributors
Dr. Michaels enjoys investigating phenotypic variations among plants, determining whether they have a genetic basis, and using them to select for improved cultivars. His current work focuses on dry edible beans, industrial hemp, sweet sorghum, and lettuce for organic and small farm production systems. He is passionate about developing and delivering effective undergraduate learning experiences for his students in face-to-face and online formats.
Dr. Hoover’s research examines production methodologies for producing fruit crops with sustainable methods, emphasizing practices that are gentle on the environment. She and her colleagues have developed systems for minimizing weed pressure in June- bearing strawberries and for producing day-neutral strawberries in cold climates. She is also part of the international research group NC140 , which studies the winter hardiness of apple rootstocks. She was appointed Head of the Department of Horticultural Science at the University of Minnesota (UMN) in 2009, and leads a diverse group of faculty and staff who work to produce knowledge on a wide range of plant species, including traditional horticultural plants, fruits, vegetables, and flowers
Ms. Irish received her BS in Horticulture, with an emphasis in public horticulture, in 2015. She went directly into her master’s program, working on a collaboration between the Iowa SNAP-Ed and Master Gardener programs that involved working with master gardeners on food security projects across the state. As both an undergrad and a graduate student she served as a teaching assistant for the introductory horticulture course labs, the hands-on horticulture lab, and the upper-level plant propagation course. She teaches the plant propagation labs at UMN and advises the Horticulture Club.
Contribute to this Page
NTRS - NASA Technical Reports Server
Available downloads, related records.

IMAGES
COMMENTS
4.0/). Department of Pharmacy, Health and Nutritional Sciences, University of Calabria, Rende, 87036 Cosenza, Italy; [email protected]. Medicinal plants represent the most ancient form ...
PDF | Plants are one of the two major groups of living organisms that are an essential entity to the function of the biosphere. Plants can be found in... | Find, read and cite all the research you ...
PDF | Growth is a widely used term in plant science and ecology, but it can have different meanings depending on the context and the spatiotemporal... | Find, read and cite all the research you ...
The Rigveda (5000 BC) has recorded 67 medicinal plants, the Yajurveda 81 species, the Atharvaveda (4500-2500 BC) 290 species, and the Charak Samhita (700 BC) and Sushrut Samhita (200 BC) have described properties and uses of 1100 and 1270 species, respectively, to compound the drugs and use.
Figure 5. Temporal evolution on medical plants publications for Top 12 countries. The first group is the leaders of this research, China and India, with between 800 and 1100 publications per year. China led the research from 1996 to 2010, and from this year to 2016, the leader was India, after which it returned to China.
Here, we investigate the global distribution of 35,687 utilized plant species spanning 10 use categories (e.g., food, medicine, material). Our findings indicate general concordance between utilized and total plant diversity, supporting the potential for simultaneously conserving species diversity and its contributions to people. Although ...
Medicinal Plants, The Governmental Conference Centre, Ottawa, Canada, 16-19 July, 2001 370 Annex 2 Cumulative index (in alphabetical order of plant name) 373 Annex 3 Cumulative index (in alphabetical order of plant material of interest) 375 Contents SMPvol3 layout.indd iv 10.8.2007 12:09:50
Plants are one of the two major groups of living organisms that are an essential entity to the function of the biosphere. Plants can be found in all known parts of the earth, in all shapes and sizes. They include the green algae, mosses, ferns, vines, grasses, bushes, herbs, flowering plants and trees. Although some plants are parasitic, most produce their own food through photosynthesis. Most ...
apter 1Medicinal Plants and Herbal Drugs: An O. rv. wBurhan Ahad, Waseem Shahri, Humeera Raso. Sumaiyah Rasool, and Tufail Hussain1.1 IntroductionPlants apart from being a source. f food are also sources of non-food industrial products. Such crops are typically cultivated on a large scale fo. the production of fine chemicals or specialty ...
Sunflower ( Helianthus annuus L.), in the Asteraceae and the Helianthus genus, is a horticultural crop with important economic and ornamental value and a major research focus. In May 2017, a high ...
2.1. Eligibility Criteria. The eligibility criteria for inclusion of a study in this research were as follows: (1) participants of any type were recruited; (2) no criteria were set for the type of indoor plant to be used in interventions; (3) the comparator was participants in an indoor environment without any plants or with other elements; (4) the outcome included any type of objectively ...
PDF | An overview of plant biodiversity is provided in this chapter. Details of genetic diversity, species diversity and ecosystem diversity are given.... | Find, read and cite all the research ...
The success of Tournefort's system resulted from the ease to identify groups of plants based on the number and relative symmetry of the petals of a flower. Within his system, Tournefort precisely defined 698 enti-ties—'Institutiones rei herbariae,' 1700—each being called a genus, plural: genera.
Fourth Edition. This latest edition of The Physiology of Flowering Plants has been completely updated to cover the explosion of interest in plant biology. A whole-plant approach has been used to produce an integrated view of plant function, covering both the fundamentals of whole plant physiology and the latest developments in molecular biology.
An approachable guide to the fundamentals of plant science. Created for horticulture students, gardeners, science teachers, and anyone interested in understanding plants and how they grow. This is the required text for HORT 1001/6001 Plant Propagation at the University of Minnesota Department of Horticultural Science.
A new way of naming plants, using only two words, s developed by Caspar Bauhin (1560-1624). August Rivinus (1652-1723) also proposed that plants ought to have names of no more than two words. You probably thought it invented by Carolus Linnaeus (1707-1778), because so many textbooks inco rectly give him the credit.
PDF | On Aug 4, 2019, Ali Haider Mohammed published Importance of Medicinal Plants | Find, read and cite all the research you need on ResearchGate
Plant taxonomy: The scientific classification of plants . We live from plants and of plants. We eat them, we use them for construction, we weave clothes with their fibers and we ferment them to make beverages. Through the process of photosynthesis, plants capture sunlight and atmospheric CO. 2 to make organic molecules. Think about it: plants ...
Interior Landscape Plants for Indoor Air Pollution Abatement In this study, the leaves, roots, soil, and associated microorganisms of plants have been evaluated as a possible means of reducing indoor air pollutants. Additionally, a novel approach of using plant systems for removing high concentrations of indoor air pollutants such as cigarette smoke, organic solvents, and possibly radon has ...
In the third edition of her successful textbook, Paula Rudall provides a comprehensive yet succinct introduction to the anatomy of flowering plants. Thoroughly revised and updated throughout, the book covers all aspects of comparative plant structure and development, arranged in a series of chapters on the stem, root, leaf, flower, seed and fruit.
Handbook for Plant Science Research: Methods, Techniques, and Essential Tools. July 2023. July 2023. ISBN: ISBN 9789392572159. Authors: Shraddha Bhaskar Sawant. Bihar Agriculture University ...
GLOBE® 2017 Plant-A-Plant: Teacher Guide - 3 Biosphere Appendix Welcome Introduction Protocols Learning Activities ENGAGE Class Time: VariesGrouping: • Students Observe Natural Phenomenon (seed germination and growth): Use the Seed Germination Laboratory Guide and Data Sheet to set up seeds for ob- servation. Every day (for 7 days, or until seedlings have 2 leaves) different students record ...
The introduction to plant nutrition addresses basic and general topics on the impor-. tance of this area to meet nutritional requirements and promote crop growth, devel-. opment, and yield. W e ...